Peptide Mimics of the Ribosomal P Stalk Inhibit the Activity of Ricin A Chain by Preventing Ribosome Binding
Abstract
:1. Introduction
2. Results
2.1. The Longer Peptides Have Higher Affinity for RTA than the Shorter Peptides
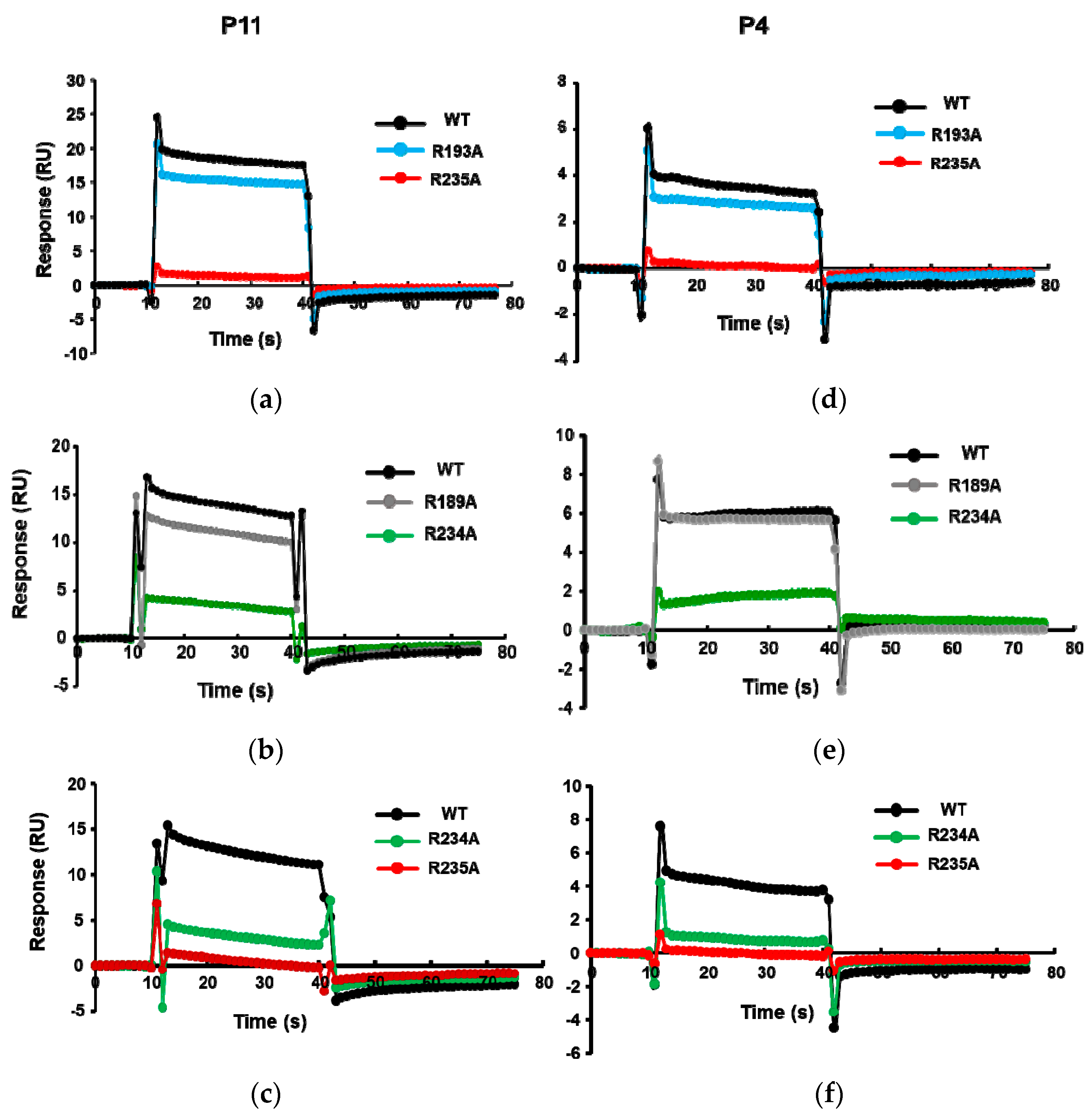
2.2. Peptides Bind to the Ribosome Binding Site of RTA
2.3. Peptides Compete with Ribosomes for Binding to RTA
2.4. Peptides Inhibit the Depurination Activity of RTA
3. Discussion
4. Materials and Methods
4.1. Peptide Synthesis
4.2. RTA Purification
4.3. Peptide–RTA Interaction
4.4. Peptide Competition Assays
4.5. Inhibition of RTA Depurination
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bolognesi, A.; Bortolotti, M.; Maiello, S.; Battelli, M.G.; Polito, L. Ribosome-inactivating proteins from plants: A historical overview. Molecules 2016, 21, 1627. [Google Scholar] [CrossRef] [PubMed]
- Olsnes, S. The history of ricin, abrin and related toxins. Toxicon 2004, 44, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Stirpe, F. Ribosome-inactivating proteins. Toxicon 2004, 44, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Tesh, V.L.; O’Brien, A.D. The pathogenic mechanisms of Shiga toxin and the Shiga-like toxins. Mol. Microbiol. 1991, 5, 1817–1822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiou, J.C.; Li, X.P.; Remacha, M.; Ballesta, J.P.; Tumer, N.E. The ribosomal stalk is required for ribosome binding, depurination of the rrna and cytotoxicity of ricin A chain in saccharomyces cerevisiae. Mol. Microbiol. 2008, 70, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- McCluskey, A.J.; Poon, G.M.; Bolewska-Pedyczak, E.; Srikumar, T.; Jeram, S.M.; Raught, B.; Gariepy, J. The catalytic subunit of Shiga-like toxin 1 interacts with ribosomal stalk proteins and is inhibited by their conserved C-terminal domain. J. Mol. Biol. 2008, 378, 375–386. [Google Scholar] [CrossRef] [PubMed]
- McCluskey, A.J.; Bolewska-Pedyczak, E.; Jarvik, N.; Chen, G.; Sidhu, S.S.; Gariepy, J. Charged and hydrophobic surfaces on the a chain of Shiga-like toxin 1 recognize the C-terminal domain of ribosomal stalk proteins. PLoS ONE 2012, 7, e31191. [Google Scholar] [CrossRef] [PubMed]
- Chiou, J.C.; Li, X.P.; Remacha, M.; Ballesta, J.P.; Tumer, N.E. Shiga toxin 1 is more dependent on the P proteins of the ribosomal stalk for depurination activity than Shiga toxin 2. Int. J. Biochem. Cell Biol. 2011, 43, 1792–1801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, W.W.; Mak, A.N.; Wong, K.B.; Shaw, P.C. Structures and ribosomal interaction of ribosome-inactivating proteins. Molecules 2016, 21, 1588. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.K.; Wong, E.C.; Lee, K.M.; Wong, K.B. Structures of eukaryotic ribosomal stalk proteins and its complex with trichosanthin, and their implications in recruiting ribosome-inactivating proteins to the ribosomes. Toxins 2015, 7, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Tumer, N.E.; Li, X.P. Interaction of ricin and Shiga toxins with ribosomes. Curr. Top. Microbiol. Immunol. 2012, 357, 1–18. [Google Scholar] [PubMed]
- Jose, M.P.; Santana-Roman, H.; Remacha, M.; Ballesta, J.P.; Zinker, S. Eukaryotic acidic phosphoproteins interact with the ribosome through their amino-terminal domain. Biochemistry 1995, 34, 7941–7948. [Google Scholar] [CrossRef] [PubMed]
- Tchorzewski, M. The acidic ribosomal P proteins. Int. J. Biochem. Cell Biol. 2002, 34, 911–915. [Google Scholar] [CrossRef]
- Krokowski, D.; Boguszewska, A.; Abramczyk, D.; Liljas, A.; Tchorzewski, M.; Grankowski, N. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol. Microbiol. 2006, 60, 386–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahl, M.C.; Moller, W. Structure and function of the acidic ribosomal stalk proteins. Curr. Protein Pept. Sci. 2002, 3, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Yusa, K.; Chu, L.O.; Yu, C.W.; Oono, M.; Miyoshi, T.; Ito, K.; Shaw, P.C.; Wong, K.B.; Uchiumi, T. Solution structure of human P1*P2 heterodimer provides insights into the role of eukaryotic stalk in recruiting the ribosome-inactivating protein trichosanthin to the ribosome. Nucleic Acids Res. 2013, 41, 8776–8787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villa, E.; Sengupta, J.; Trabuco, L.G.; Lebarron, J.; Baxter, W.T.; Shaikh, T.R.; Grassucci, R.A.; Nissen, P.; Ehrenberg, M.; Schulten, K.; et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc. Natl. Acad. Sci. USA 2009, 106, 1063–1068. [Google Scholar] [CrossRef] [PubMed]
- Schmeing, T.M.; Voorhees, R.M.; Kelley, A.C.; Gao, Y.G.; Murphy, F.V.T.; Weir, J.R.; Ramakrishnan, V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 2009, 326, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Tanzawa, T.; Kato, K.; Girodat, D.; Ose, T.; Kumakura, Y.; Wieden, H.J.; Uchiumi, T.; Tanaka, I.; Yao, M. The C-terminal helix of ribosomal P stalk recognizes a hydrophobic groove of elongation factor 2 in a novel fashion. Nucleic Acids Res. 2018, 46, 3232–3244. [Google Scholar] [CrossRef] [PubMed]
- Murakami, R.; Singh, C.R.; Morris, J.; Tang, L.; Harmon, I.; Azuma, T.; Miyoshi, T.; Ito, K.; Asano, K.; Uchiumi, T. The interaction between the ribosomal stalk proteins and translation initiation factor 5B promotes translation initiation. Mol. Cell. Biol. 2018, 38, e00067-18. [Google Scholar] [CrossRef] [PubMed]
- May, K.L.; Li, X.P.; Martinez-Azorin, F.; Ballesta, J.P.; Grela, P.; Tchorzewski, M.; Tumer, N.E. The P1/P2 proteins of the human ribosomal stalk are required for ribosome binding and depurination by ricin in human cells. FEBS J. 2012, 279, 3925–3936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.P.; Kahn, P.C.; Kahn, J.N.; Grela, P.; Tumer, N.E. Arginine residues on the opposite side of the active site stimulate the catalysis of ribosome depurination by ricin A chain by interacting with the P-protein stalk. J. Biol. Chem. 2013, 288, 30270–30284. [Google Scholar] [CrossRef] [PubMed]
- Li, X.P.; Chiou, J.C.; Remacha, M.; Ballesta, J.P.; Tumer, N.E. A two-step binding model proposed for the electrostatic interactions of ricin A chain with ribosomes. Biochemistry 2009, 48, 3853–3863. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Link, T.M.; Schramm, V.L. Ricin A-chain: Kinetics, mechanism, and RNA stem-loop inhibitors. Biochemistry 1998, 37, 11605–11613. [Google Scholar] [CrossRef] [PubMed]
- Krokowski, D.; Tchorzewski, M.; Boguszewska, A.; Grankowski, N. Acquisition of a stable structure by yeast ribosomal P0 protein requires binding of P1a-P2b complex: In vitro formation of the stalk structure. Biochim. Biophys. Acta 2005, 1724, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Li, X.P.; Tumer, N.E. Differences in ribosome binding and sarcin/ricin loop depurination by Shiga and ricin holotoxins. Toxins 2017, 9, 133. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.W.; Tang, Y.S.; Sze, S.Y.; Zhu, Z.N.; Wong, K.B.; Shaw, P.C. Crystal structure of ribosome-inactivating protein ricin A chain in complex with the C-terminal peptide of the ribosomal stalk protein P2. Toxins 2016, 8, 296. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhu, Y.; Wang, C.; Niu, L.; Teng, M.; Li, X. Structural insights into the interaction of the ribosomal P stalk protein P2 with a type II ribosome-inactivating protein ricin. Sci. Rep. 2016, 6, 37803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Li, X.P.; Chen, B.Y.; Tumer, N.E. Ricin uses arginine 235 as an anchor residue to bind to P-proteins of the ribosomal stalk. Sci. Rep. 2017, 7, 42912. [Google Scholar] [CrossRef] [PubMed]
- Sturm, M.B.; Schramm, V.L. Detecting ricin: Sensitive luminescent assay for ricin A-chain ribosome depurination kinetics. Anal. Chem. 2009, 81, 2847–2853. [Google Scholar] [CrossRef] [PubMed]
- Pierce, M.; Kahn, J.N.; Chiou, J.; Tumer, N.E. Development of a quantitative RT-PCR assay to examine the kinetics of ribosome depurination by ribosome inactivating proteins using Saccharomyces cerevisiae as a model. RNA 2011, 17, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Wahome, P.G.; Robertus, J.D.; Mantis, N.J. Small-molecule inhibitors of ricin and Shiga toxins. Curr. Top. Microbiol. 2012, 357, 179–207. [Google Scholar]
- Stechmann, B.; Bai, S.K.; Gobbo, E.; Lopez, R.; Merer, G.; Pinchard, S.; Panigai, L.; Tenza, D.; Raposo, G.; Beaumelle, B.; et al. Inhibition of retrograde transport protects mice from lethal ricin challenge. Cell 2010, 141, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.S.; Chu, L.O.; Lee, K.M.; Too, P.H.; Ma, K.W.; Sze, K.H.; Zhu, G.; Shaw, P.C.; Wong, K.B. Interaction between trichosanthin, a ribosome-inactivating protein, and the ribosomal stalk protein P2 by chemical shift perturbation and mutagenesis analyses. Nucleic Acids Res. 2007, 35, 1660–1672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Too, P.H.M.; Ma, M.K.W.; Mak, A.N.S.; Wong, Y.T.; Tung, C.K.C.; Zhu, G.; Au, S.W.N.; Wong, K.B.; Shaw, P.C. The C-terminal fragment of the ribosomal P protein complexed to trichosanthin reveals the interaction between the ribosome-inactivating protein and the ribosome. Nucleic Acids Res. 2009, 37, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Li, X.-P.; Kahn, J.N.; Tumer, N.E. Functional assays for measuring the catalytic activity of ribosome inactivating proteins. Toxins 2018, 10, 240. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Honda, T.; Suzuki, T.; Miyoshi, T.; Murakami, R.; Yao, M.; Uchiumi, T. Molecular insights into the interaction of the ribosomal stalk protein with elongation factor 1α. Nucleic Acids Res. 2014, 42, 14042–14052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nomura, N.; Honda, T.; Baba, K.; Naganuma, T.; Tanzawa, T.; Arisaka, F.; Noda, M.; Uchiyama, S.; Tanaka, I.; Yao, M.; et al. Archaeal ribosomal stalk protein interacts with translation factors in a nucleotide-independent manner via its conserved C terminus. Proc. Natl. Acad. Sci. USA 2012, 109, 3748–3753. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Peptides | Sequence | MW | KD (µM) * |
|---|---|---|---|
| P11 | S105DDDMGFGLFD115 | 1218.25 | 196 ± 17 |
| P10 | D106DDMGFGLFD115 | 1131.18 | 272 ± 6 |
| P9 | D107DMGFGLFD115 | 1016.09 | 309 ± 7 |
| P8 | D108MGFGLFD115 | 901.00 | 299 ± 5 |
| P7 | M109GFGLFD115 | 785.91 | 294 ± 47 |
| P6 | G110FGLFD115 | 654.71 | 399 ± 20 |
| P5 | F111GLFD115 | 597.66 | 497 ± 30 |
| P4 | G112LFD115 | 450.49 | 451 ± 17 |
| P3 | L113FD115 | 393.44 | >10 mM |
| Peptide | Sequence | Yeast Ribosome IC50 (µM) * | Rat Ribosome IC50 (µM) * |
|---|---|---|---|
| P11 | S105DDDMGFGLFD115 | 4.7 ± 0.9 | 31 ± 6.3 |
| P10 | D106DDMGFGLFD115 | 7.9 ± 1.7 | 83 ± 18 |
| P9 | D107DMGFGLFD115 | 15 ± 1.5 | 142 ± 61 |
| P8 | D108MGFGLFD115 | 23 ± 4.4 | 267 ± 80 |
| P7 | M109GFGLFD115 | 34 ± 9.5 | NA |
| P6 | G110FGLFD115 | 63 ± 13 | NA |
| P5 | F111GLFD115 | 121 ± 44 | NA |
| P4 | G112LFD115 | 102 ± 45 | NA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.-P.; Kahn, J.N.; Tumer, N.E. Peptide Mimics of the Ribosomal P Stalk Inhibit the Activity of Ricin A Chain by Preventing Ribosome Binding. Toxins 2018, 10, 371. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10090371
Li X-P, Kahn JN, Tumer NE. Peptide Mimics of the Ribosomal P Stalk Inhibit the Activity of Ricin A Chain by Preventing Ribosome Binding. Toxins. 2018; 10(9):371. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10090371
Chicago/Turabian StyleLi, Xiao-Ping, Jennifer N. Kahn, and Nilgun E. Tumer. 2018. "Peptide Mimics of the Ribosomal P Stalk Inhibit the Activity of Ricin A Chain by Preventing Ribosome Binding" Toxins 10, no. 9: 371. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10090371




