Two Clostridium perfringens Type E Isolates in France
Abstract
:1. Introduction
2. Results
2.1. Identification of Two C. perfringens Type E and Investigation of Iota Toxin Production
2.2. In Vitro Actin ADP-Ribosylation
2.3. Whole Genome Sequencing
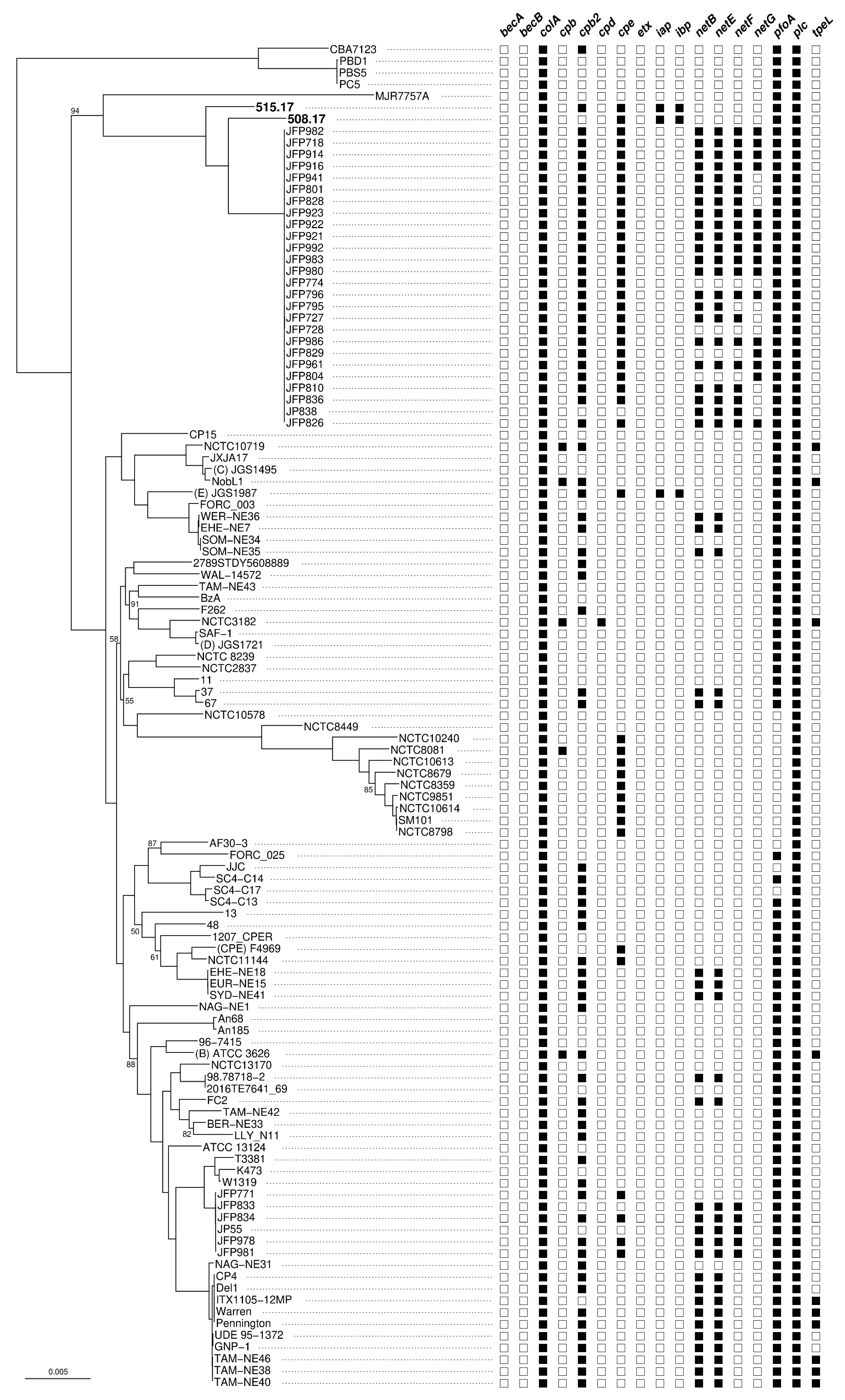
2.4. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. C. perfringens Isolates
4.2. C. perfringens Cultures
4.3. Iota Toxin Production and Purification
4.4. Western Blotting
4.5. ADP-Ribosylation
4.6. Cytotoxicity Assay
4.7. Genome Sequencing, Assembly, and Analysis
4.8. Phylogenetic Reconstruction
4.9. Sequence Accession
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Petit, L.; Gibert, M.; Popoff, M.R. Clostridium perfringens: Toxinotype and genotype. Trends Microbiol. 1999, 7, 104–110. [Google Scholar] [CrossRef]
- Rood, J.I.; Adams, V.; Lacey, J.; Lyras, D.; McClane, B.A.; Melville, S.B.; Moore, R.J.; Popoff, M.R.; Sarker, M.R.; Songer, J.G.; et al. Expansion of the Clostridium perfringens toxin-based typing scheme. Anaerobe 2018, 53, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Stiles, B.G.; Pradhan, K.; Fleming, J.M.; Samy, R.P.; Barth, H.; Popoff, M.R. Clostridium and Bacillus binary enterotoxins: Bad for the bowels, and eukaryotic being. Toxins 2014, 6, 2626–2656. [Google Scholar] [CrossRef] [PubMed]
- Barth, H.; Aktories, K.; Popoff, M.R.; Stiles, B.G. Binary bacterial toxins: Biochemistry, biology, and applications of common Clostridium and Bacillus proteins. Microbiol. Mol. Biol. Rev. 2004, 68, 373–402. [Google Scholar] [CrossRef] [PubMed]
- Songer, J.G. Clostridial diseases in domestic animals. In Handbook on Clostridia; Dürre, P., Ed.; CRC Press, Taylor and Francis Group: Boca Raton, FL, USA, 2005; pp. 527–542. [Google Scholar]
- Songer, J.G.; Miskimmins, D.W. Clostridium perfringens type E enteritis in calves: Two cases and a brief review of the literature. Anaerobe 2004, 10, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Uzal, F.A.; Songer, J.G. Diagnosis of Clostridium perfringens intestinal infections in sheep and goats. J. Vet. Diagn. Investig. 2008, 20, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Keokilwe, L.; Olivier, A.; Burger, W.P.; Joubert, H.; Venter, E.H.; Morar-Leather, D. Bacterial enteritis in ostrich (Struthio Camelus) chicks in the Western Cape Province, South Africa. Poult. Sci. 2015, 94, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Byun, J.W.; Roh, I.S.; Bae, Y.C.; Lee, M.H.; Kim, B.; Songer, J.G.; Jung, B.Y. First isolation of Clostridium perfringens type E from a goat with diarrhea. Anaerobe 2013, 22, 141–143. [Google Scholar] [CrossRef] [PubMed]
- Redondo, L.M.; Farber, M.; Venzano, A.; Jost, B.H.; Parma, Y.R.; Fernandez-Miyakawa, M.E. Sudden death syndrome in adult cows associated with Clostridium perfringens type E. Anaerobe 2013, 20, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Songer, J.G. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 1996, 9, 216–234. [Google Scholar] [CrossRef] [PubMed]
- Ferrarezi, M.C.; Cardoso, T.C.; Dutra, I.S. Genotyping of Clostridium perfringens isolated from calves with neonatal diarrhea. Anaerobe 2008, 14, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Billington, S.J.; Wieckowski, E.U.; Sarker, M.R.; Bueschel, D.; Songer, J.G.; McClane, B.A. Clostridium perfringens type E animal enteritis isolates with highly conserved, silent enterotoxin gene sequences. Infect. Immun. 1998, 66, 4531–4536. [Google Scholar] [PubMed]
- Popoff, M.R. Detection of toxigenic Clostridia. In PCR Detection of Microbial Pathogens; Sachse, K., Frey, J., Eds.; Humana Press: Totowa, NJ, USA, 2002; Volume 216, pp. 137–152. [Google Scholar]
- Irikura, D.; Monma, C.; Suzuki, Y.; Nakama, A.; Kai, A.; Fukui-Miyazaki, A.; Horiguchi, Y.; Yoshinari, T.; Sugita-Konishi, Y.; Kamata, Y. Identification and Characterization of a New Enterotoxin Produced by Clostridium perfringens Isolated from Food Poisoning Outbreaks. PLoS ONE 2015, 10, e0138183. [Google Scholar] [CrossRef] [PubMed]
- Yonogi, S.; Matsuda, S.; Kawai, T.; Yoda, T.; Harada, T.; Kumeda, Y.; Gotoh, K.; Hiyoshi, H.; Nakamura, S.; Kodama, T.; et al. BEC, a novel enterotoxin of Clostridium perfringens found in human clinical isolates from acute gastroenteritis outbreaks. Infect. Immun. 2014, 82, 2390–2399. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Miyamoto, K.; McClane, B.A. Comparison of virulence plasmids among Clostridium perfringens type E isolates. Infect. Immun. 2007, 75, 1811–1819. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Yumine, N.; Mimura, K.; Nagahama, M.; Li, J.; McClane, B.A.; Akimoto, S. Identification of novel Clostridium perfringens type E strains that carry an iota toxin plasmid with a functional enterotoxin gene. PLoS ONE 2011, 6, e20376. [Google Scholar] [CrossRef] [PubMed]
- Perelle, S.; Gibert, M.; Boquet, P.; Popoff, M.R. Characterization of Clostridium perfringens iota-toxin genes and expression in Escherichia coli. Infect. Immun. 1993, 61, 5147–5156. [Google Scholar] [PubMed]
- Toniti, W.; Yoshida, T.; Tsurumura, T.; Irikura, D.; Monma, C.; Kamata, Y.; Tsuge, H. Crystal structure and structure-based mutagenesis of actin-specific ADP-ribosylating toxin CPILE-a as novel enterotoxin. PLoS ONE 2017, 12, e0171278. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, A.; Miyamoto, K.; Kuwahara, T.; Miki, Y.; Kaneko, I.; Li, J.; McClane, B.A.; Akimoto, S. Genetic characterization of type A enterotoxigenic Clostridium perfringens strains. PLoS ONE 2009, 4, e5598. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Wagendorp, A.; Moezelaar, R.; Abee, T.; Wells-Bennik, M.H. A wide variety of Clostridium perfringens type A food-borne isolates that carry a chromosomal cpe gene belong to one multilocus sequence typing cluster. Appl. Environ. Microbiol. 2012, 78, 7060–7068. [Google Scholar] [CrossRef] [PubMed]
- Gibert, M.; Petit, L.; Raffestin, S.; Okabe, A.; Popoff, M.R. Clostridium perfringens iota-toxin requires activation of both binding and enzymatic components for cytopathic activity. Infect. Immun. 2000, 68, 3848–3853. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Popoff, M.R.; Milward, F.W.; Bancillon, B.; Boquet, P. Purification of the Clostridium spiroforme binary toxin and activity of the toxin on HEp-2 cells. Infect. Immun. 1989, 57, 2462–2469. [Google Scholar] [PubMed]
- Perelle, S.; Scalzo, S.; Kochi, S.; Mock, M.; Popoff, M.R. Immunological and functional comparison between Clostridium perfringens iota toxin, C. spiroforme toxin, and anthrax toxins. FEMS Microbiol. Lett. 1997, 146, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Mazuet, C.; Legeay, C.; Sautereau, J.; Ma, L.; Bouchier, C.; Bouvet, P.; Popoff, M.R. Diversity of Group I and II Clostridium botulinum strains from France including recently Identified subtypes. Genome Biol. Evol. 2016, 8, 1643–1660. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Seeman, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Galardini, M.; Biondi, E.G.; Bazzicalupo, M.; Mengoni, A. CONTIGuator: A bacterial genomes finishing tool for structural insights on draft genomes. Source Code Biol. Med. 2011, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Ohtani, K.; Hirakawa, H.; Ohshima, K.; Yamashita, A.; Shiba, T.; Ogasawara, N.; Hattori, M.; Kuhara, S.; Hayashi, H. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 2002, 99, 996–1001. [Google Scholar] [CrossRef] [PubMed]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]


| Strain/Sequence | Iota Ia Similarity % (aa) | Iota Ib Similarity % (aa) | ||||||
|---|---|---|---|---|---|---|---|---|
| 508.17 | pCPPB-1 | 515.17 | X73562 | 508.17 | pCPPB-1 | 515.17 | X73562 | |
| 508.17 | 100.00 | 100.00 | ||||||
| pCPPB-1 | 99.87 | 100.00 | 99.85 | 100.00 | ||||
| 515.17 | 99.73 | 99.86 | 100.00 | 99.93 | 99.86 | 100.00 | ||
| X73562 | 91.20 | 91.33 | 91.19 | 100.00 | 89.69 | 89.77 | 89.77 | 100.00 |
| Strain/Sequence | Variable aa Positions on cpe | Similarity % | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18 | 20 | 32 | 172 | 193 | 217 | 257 | 275 | 276 | 283 | 287 | 313 | 508.17 | 515.17 | pCPPB-1 | pCPF4969 | |
| 508.17 | L | V | K | A | S | T | R | Q | E | N | I | A | - | |||
| 515.17 | L | V | K | A | S | T | R | Q | E | N | I | A | 99.07 | - | ||
| pCPPB-1 | L | V | K | A | S | T | R | Q | E | N | I | A | 100.00 | 99.07 | - | |
| pCPF4969 | F | I | N | G | T | S | K | E | Q | K | V | S | 96.36 | 97.3 | 96.36 | - |
| pJFP838-all | F | I | N | G | T | S | K | E | Q | K | V | S | 96.36 | 97.3 | 96.36 | 100.00 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diancourt, L.; Sautereau, J.; Criscuolo, A.; Popoff, M.R. Two Clostridium perfringens Type E Isolates in France. Toxins 2019, 11, 138. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11030138
Diancourt L, Sautereau J, Criscuolo A, Popoff MR. Two Clostridium perfringens Type E Isolates in France. Toxins. 2019; 11(3):138. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11030138
Chicago/Turabian StyleDiancourt, Laure, Jean Sautereau, Alexis Criscuolo, and Michel R. Popoff. 2019. "Two Clostridium perfringens Type E Isolates in France" Toxins 11, no. 3: 138. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11030138





