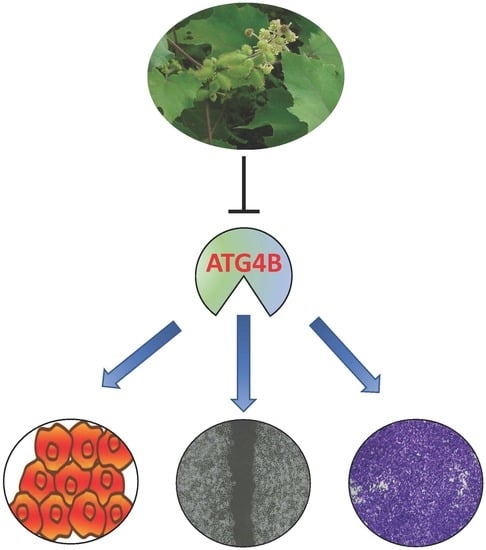
Xanthium strumarium Fruit Extract Inhibits ATG4B and Diminishes the Proliferation and Metastatic Characteristics of Colorectal Cancer Cells
Abstract
:1. Introduction
2. Results
2.1. Screening Plant Extracts for Potential ATG4B Inhibitors
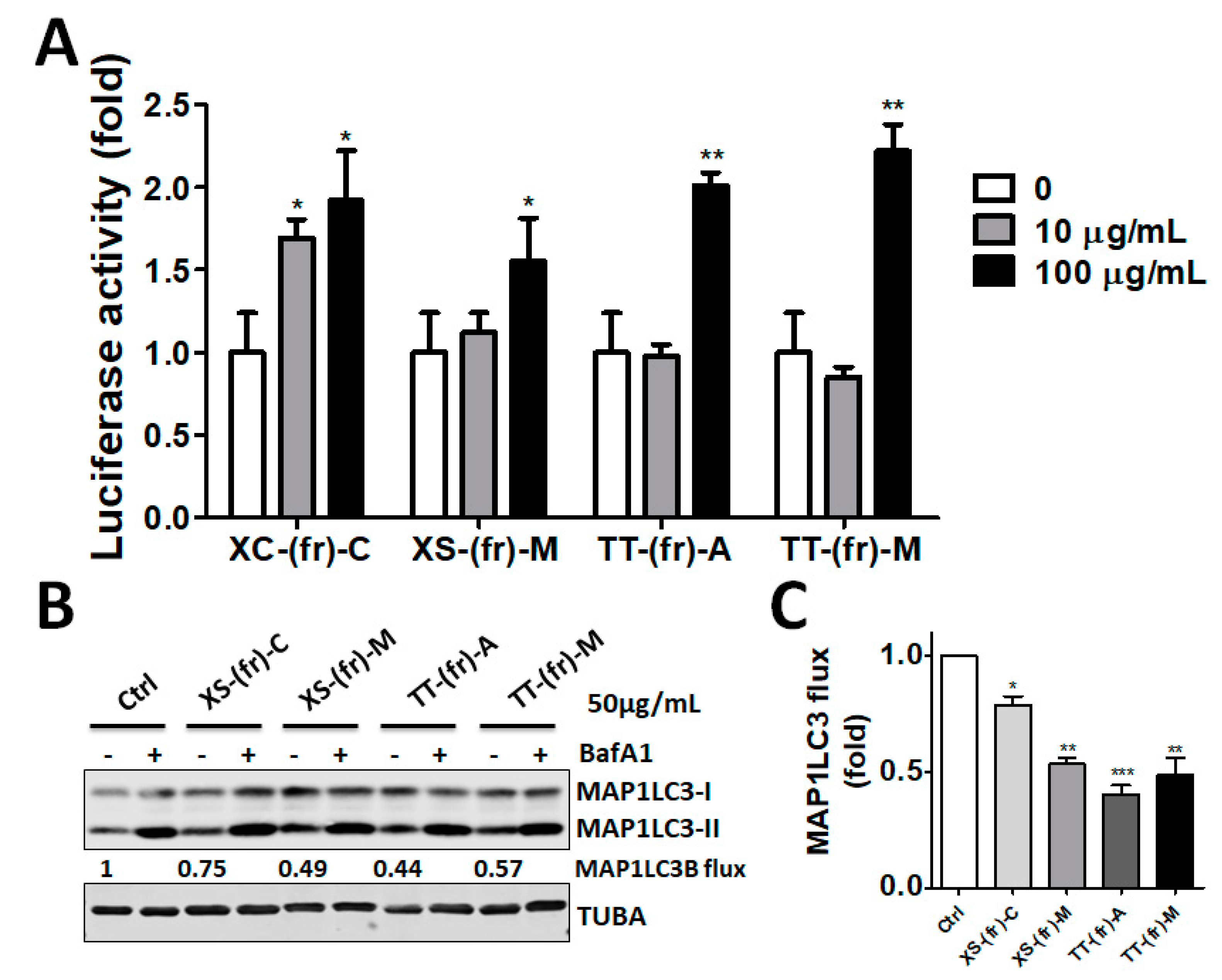
2.2. Validation of the Effect of the Plant Extracts on Cellular ATG4B Activity and Autophagic Activity
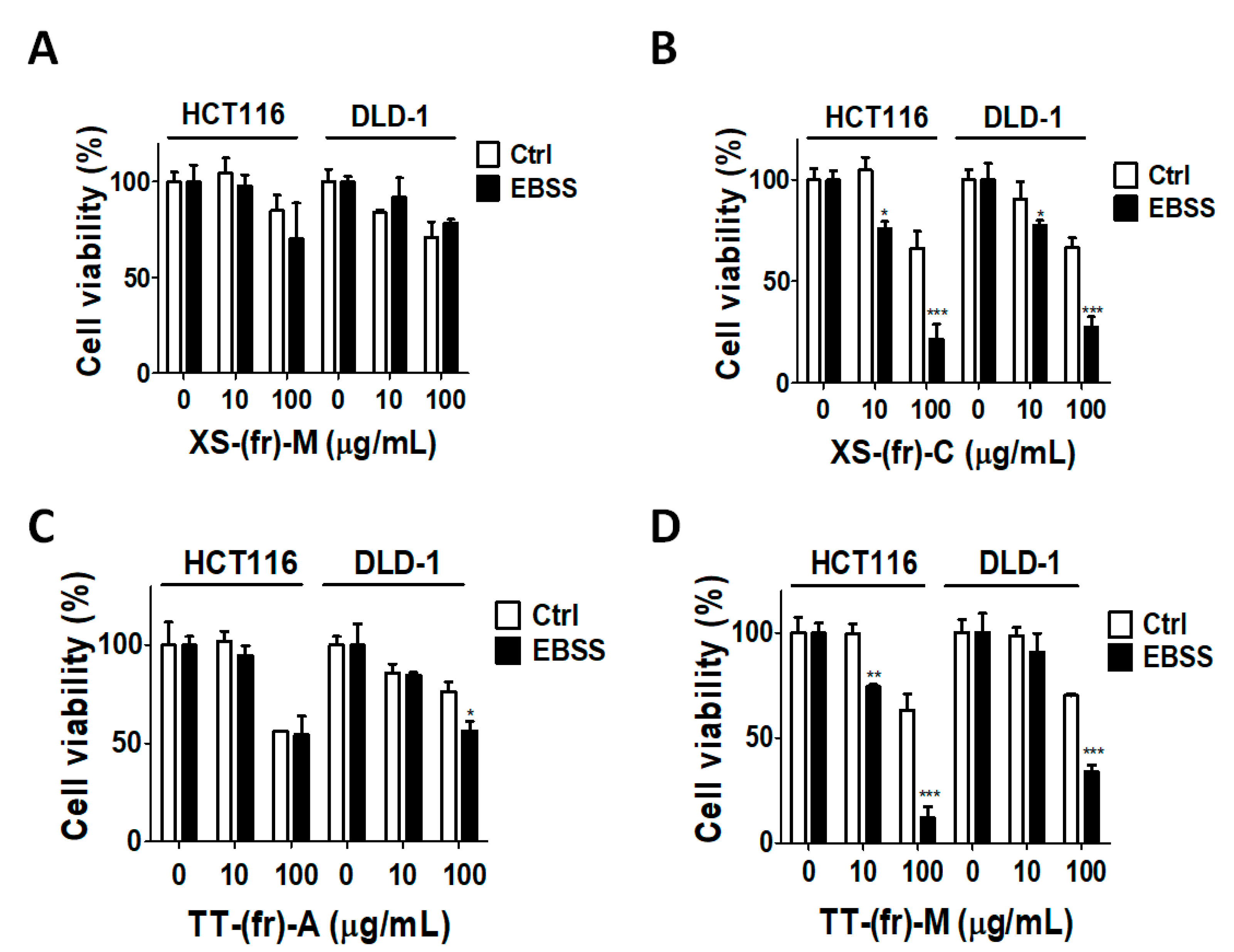
2.3. Effects of Plant Extracts on Colorectal Cancer Cells
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plasmid Construction
5.2. Plant Extract Isolation
5.3. Protein Purification and Activity Assay
5.4. Autophagic Flux and Immunoblotting
5.5. Cell Viability Assay
5.6. Wound-Healing Assay
5.7. Cell Invasion Assays
5.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Amaravadi, R.; Kimmelman, A.C.; White, E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016, 30, 1913–1930. [Google Scholar] [CrossRef] [PubMed]
- Takamura, A.; Komatsu, M.; Hara, T.; Sakamoto, A.; Kishi, C.; Waguri, S.; Eishi, Y.; Hino, O.; Tanaka, K.; Mizushima, N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011, 25, 795–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.M.; Jiang, Z.F.; Ding, P.S.; Shao, L.J.; Liu, R.Y. Hypoxia-induced autophagy mediates cisplatin resistance in lung cancer cells. Sci. Rep. 2015, 5, 12291. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yin, H.; Zhang, Y.; Li, X.; Tong, H.; Zeng, Y.; Wang, Q.; He, W. Hypoxia-induced autophagy promotes gemcitabine resistance in human bladder cancer cells through hypoxia-inducible factor 1alpha activation. Int. J. Oncol. 2018, 53, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.F.; Tsai, K.L.; Hsu, C.J.; Tsai, W.L.; Cheng, J.S.; Chang, H.W.; Shiau, C.W.; Goan, Y.G.; Tseng, H.H.; Wu, C.H.; et al. Drug Repurposing Screening Identifies Tioconazole as an ATG4 Inhibitor that Suppresses Autophagy and Sensitizes Cancer Cells to Chemotherapy. Theranostics 2018, 8, 830–845. [Google Scholar] [CrossRef]
- Chude, C.I.; Amaravadi, R.K. Targeting Autophagy in Cancer: Update on Clinical Trials and Novel Inhibitors. Int. J. Mol. Sci. 2017, 18, 1279. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Hou, Y.; Wang, J.; Chen, X.; Shao, Z.M.; Yin, X.M. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J. Biol. Chem. 2011, 286, 7327–7338. [Google Scholar] [CrossRef]
- Shu, C.-W.; Drag, M.; Bekes, M.; Zhai, D.; Salvesen, G.S.; Reed, J.C. Synthetic substrates for measuring activity of autophagy proteases: Autophagins (Atg4). Autophagy 2010, 6, 936–947. [Google Scholar] [CrossRef]
- Liu, P.F.; Leung, C.M.; Chang, Y.H.; Cheng, J.S.; Chen, J.J.; Weng, C.J.; Tsai, K.W.; Hsu, C.J.; Liu, Y.C.; Hsu, P.C.; et al. ATG4B promotes colorectal cancer growth independent of autophagic flux. Autophagy 2014, 10, 1454–1465. [Google Scholar] [CrossRef] [Green Version]
- Rothe, K.; Lin, H.; Lin, K.B.; Leung, A.; Wang, H.M.; Malekesmaeili, M.; Brinkman, R.R.; Forrest, D.L.; Gorski, S.M.; Jiang, X. The core autophagy protein ATG4B is a potential biomarker and therapeutic target in CML stem/progenitor cells. Blood 2014, 123, 3622–3634. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.F.; Hsu, C.J.; Tsai, W.L.; Cheng, J.S.; Chen, J.J.; Huang, I.F.; Tseng, H.H.; Chang, H.W.; Shu, C.W. Ablation of ATG4B Suppressed Autophagy and Activated AMPK for Cell Cycle Arrest in Cancer Cells. Cell. Physiol. Biochem. 2017, 44, 728–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bortnik, S.; Choutka, C.; Horlings, H.M.; Leung, S.; Baker, J.H.; Lebovitz, C.; Dragowska, W.H.; Go, N.E.; Bally, M.B.; Minchinton, A.I.; et al. Identification of breast cancer cell subtypes sensitive to ATG4B inhibition. Oncotarget 2016, 7, 66970–66988. [Google Scholar] [CrossRef] [PubMed]
- Toshima, T.; Shirabe, K.; Matsumoto, Y.; Yoshiya, S.; Ikegami, T.; Yoshizumi, T.; Soejima, Y.; Ikeda, T.; Maehara, Y. Autophagy enhances hepatocellular carcinoma progression by activation of mitochondrial beta-oxidation. J. Gastroenterol. 2014, 49, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Bosc, D.; Vezenkov, L.; Bortnik, S.; An, J.; Xu, J.; Choutka, C.; Hannigan, A.M.; Kovacic, S.; Loo, S.; Clark, P.G.K.; et al. A new quinoline-based chemical probe inhibits the autophagy-related cysteine protease ATG4B. Sci. Rep. 2018, 8, 11653. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.W.; Madiraju, C.; Zhai, D.; Welsh, K.; Diaz, P.; Sergienko, E.; Sano, R.; Reed, J.C. High-throughput fluorescence assay for small-molecule inhibitors of autophagins/Atg4. J. Biomol. Screen. 2011, 16, 174–182. [Google Scholar] [CrossRef]
- Xu, D.; Xu, Z.; Han, L.; Liu, C.; Zhou, Z.; Qiu, Z.; Lin, X.; Tang, G.; Shen, H.; Aebi, J.; et al. Identification of New ATG4B Inhibitors Based on a Novel High-Throughput Screening Platform. SLAS Discov. 2017, 22, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Gali-Muhtasib, H.; Hmadi, R.; Kareh, M.; Tohme, R.; Darwiche, N. Cell death mechanisms of plant-derived anticancer drugs: Beyond apoptosis. Apoptosis 2015, 20, 1531–1562. [Google Scholar] [CrossRef]
- Catanzaro, E.; Greco, G.; Potenza, L.; Calcabrini, C.; Fimognari, C. Natural Products to Fight Cancer: A Focus on Juglans regia. Toxins 2018, 10, 469. [Google Scholar] [CrossRef]
- Pan, S.Y.; Zhou, S.F.; Gao, S.H.; Yu, Z.L.; Zhang, S.F.; Tang, M.K.; Sun, J.N.; Ma, D.L.; Han, Y.F.; Fong, W.F.; et al. New Perspectives on How to Discover Drugs from Herbal Medicines: CAM’s Outstanding Contribution to Modern Therapeutics. Evid. Based Complementary Altern. Med. 2013, 2013, 627375. [Google Scholar] [CrossRef]
- Giansanti, F.; Flavell, D.J.; Angelucci, F.; Fabbrini, M.S.; Ippoliti, R. Strategies to Improve the Clinical Utility of Saporin-Based Targeted Toxins. Toxins 2018, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Chen, N. Resveratrol as a Natural Autophagy Regulator for Prevention and Treatment of Alzheimer’s Disease. Nutrients 2017, 9, 927. [Google Scholar] [CrossRef]
- Shakeri, A.; Cicero, A.F.G.; Panahi, Y.; Mohajeri, M.; Sahebkar, A. Curcumin: A naturally occurring autophagy modulator. J. Cell. Physiol. 2019, 234, 5643–5654. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Yan, J.; Sun, Z. D-limonene exhibits anti-inflammatory and antioxidant properties in an ulcerative colitis rat model via regulation of iNOS, COX-2, PGE2 and ERK signaling pathways. Mol. Med. Rep. 2017, 15, 2339–2346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mowers, E.E.; Sharifi, M.N.; Macleod, K.F. Autophagy in cancer metastasis. Oncogene 2017, 36, 1619–1630. [Google Scholar] [CrossRef] [PubMed]
- Betin, V.M.; Lane, J.D. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J. Cell Sci. 2009, 122, 2554–2566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Z.; Sanchez-Lopez, E.; Karin, M. Autophagy, Inflammation, and Immunity: A Troika Governing Cancer and Its Treatment. Cell 2016, 166, 288–298. [Google Scholar] [CrossRef] [Green Version]
- Wolf, J.; Dewi, D.L.; Fredebohm, J.; Muller-Decker, K.; Flechtenmacher, C.; Hoheisel, J.D.; Boettcher, M. A mammosphere formation RNAi screen reveals that ATG4A promotes a breast cancer stem-like phenotype. Breast Cancer Res. 2013, 15, R109. [Google Scholar] [CrossRef]
- Gong, C.; Bauvy, C.; Tonelli, G.; Yue, W.; Delomenie, C.; Nicolas, V.; Zhu, Y.; Domergue, V.; Marin-Esteban, V.; Tharinger, H.; et al. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem-like/progenitor cells. Oncogene 2013, 32, 2261–2272. [Google Scholar] [CrossRef]
- Ojha, R.; Jha, V.; Singh, S.K. Gemcitabine and mitomycin induced autophagy regulates cancer stem cell pool in urothelial carcinoma cells. Biochim. Biophys. Acta 2016, 1863, 347–359. [Google Scholar] [CrossRef]
- Sun, R.; Shen, S.; Zhang, Y.J.; Xu, C.F.; Cao, Z.T.; Wen, L.P.; Wang, J. Nanoparticle-facilitated autophagy inhibition promotes the efficacy of chemotherapeutics against breast cancer stem cells. Biomaterials 2016, 103, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Evdokimova, V.; Tognon, C.; Ng, T.; Sorensen, P.H. Reduced proliferation and enhanced migration: Two sides of the same coin? Molecular mechanisms of metastatic progression by YB-1. Cell Cycle 2009, 8, 2901–2906. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, D.; Somarelli, J.A.; Hanna, G.; Palmer, G.M.; Garcia-Blanco, M.A. Cellular migration and invasion uncoupled: Increased migration is not an inexorable consequence of epithelial-to-mesenchymal transition. Mol. Cell. Biol. 2014, 34, 3486–3499. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, J.; Cao, N.; Li, Z.; Han, J.; Li, L. Resveratrol, an activator of SIRT1, induces protective autophagy in non-small-cell lung cancer via inhibiting Akt/mTOR and activating p38-MAPK. Onco Targets Ther. 2018, 11, 7777–7786. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhou, Y.; Yang, J.; Li, H.; Zhang, H.; Zheng, P. Curcumin induces apoptotic cell death and protective autophagy in human gastric cancer cells. Oncol. Rep. 2017, 37, 3459–3466. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Han, X.; Zheng, S.; Li, Z.; Sha, Y.; Ni, J.; Sun, Z.; Qiao, S.; Song, Z. Curcumin induces autophagy, inhibits proliferation and invasion by downregulating AKT/mTOR signaling pathway in human melanoma cells. Oncol. Rep. 2016, 35, 1065–1074. [Google Scholar] [CrossRef]
- Li, H.; Chen, C. Inhibition of autophagy enhances synergistic effects of Salidroside and anti-tumor agents against colorectal cancer. BMC Complement. Altern. Med. 2017, 17, 538. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.C.; Hung, W.T.; Tsai, W.L.; Lai, F.Y.; Lin, Y.S.; Huang, M.S.; Chen, J.J.; Lin, W.Y.; Weng, J.R.; Chang, T.H. Ficus septica plant extracts for treating Dengue virus in vitro. PeerJ 2017, 5, e3448. [Google Scholar] [CrossRef]




| No. | Botanical Name | Extract | Family | Part of Plant |
|---|---|---|---|---|
| 1 | Saurauia tristyla var. oldhamii | Acetone | Rubiaceae | Leaves |
| 2 | CHCl3 | Leaves | ||
| 3 | MeOH | Leaves | ||
| 4 | Fraxinus griffithii | Acetone | Oleaceae | Leaves |
| 5 | CHCl3 | Leaves | ||
| 6 | MeOH | Leaves | ||
| 7 | Phytolacca americana | Acetone | Phytolaccaceae | Whole plant |
| 8 | CHCl3 | Whole plant | ||
| 9 | MeOH | Whole plant | ||
| 10 | Elaeocarpus sylvestris | Acetone | Elaeocarpaceae | Leaves |
| 11 | CHCl3 | Leaves | ||
| 12 | MeOH | Leaves | ||
| 13 | MeOH | Stem | ||
| 14 | Piper kadsura | MeOH | Piperaceae | Stem |
| 15 | Itea parviflora | MeOH | Saxifragaceae | Stem |
| 16 | Callicarpa formosana | Acetone | Verbenaceae | Leaves |
| 17 | MeOH | Leaves | ||
| 18 | MeOH | Stem | ||
| 19 | Callicarpa kochiana | Acetone | Verbenaceae | Leaves |
| 20 | CHCl3 | Leaves | ||
| 21 | MeOH | Leaves | ||
| 22 | MeOH | Stem | ||
| 23 | MeOH | Stem | ||
| 24 | Ficus septica | Acetone | Moraceae | Leaves |
| 25 | CHCl3 | Leaves | ||
| 26 | MeOH | Leaves | ||
| 27 | MeOH | Fruit | ||
| 28 | MeOH | Heartwood | ||
| 29 | Ficus sarmentosa var. henryi | Acetone | Moraceae | Leaves |
| 30 | CHCl3 | Leaves | ||
| 31 | MeOH | Leaves | ||
| 32 | MeOH | Stem | ||
| 33 | Xanthium strumarium | CHCl3 | Asteraceae | Fruit |
| 34 | MeOH | Fruit | ||
| 35 | Tribulus terrestris | Acetone | Zygophyllaceae | Fruit |
| 36 | MeOH | Fruit | ||
| 37 | Cornus officinalis | Acetone | Cornaceae | Whole plant |
| 38 | MeOH | Whole plant | ||
| 39 | Alisma orientale | MeOH | Alismataceae | Whole plant |
| 40 | Asparagus cochinchinensis | MeOH | Asparagaceae | Root |
| 41 | Broussonetia papyrifera | MeOH | Moraceae | Leaves |
| 42 | Clausena excavata | MeOH | Rutaceae | Leaves |
| 43 | Cinnamomum insulari-montanum | MeOH | Lauraceae | Leaves |
| 44 | Lumnitzera racemosa | MeOH | Combretaceae | Leaves |
| 45 | Pueraria lobata | MeOH | Fabaceae | Whole plant |
| 46 | Sida acuta | MeOH | Malvaceae | Whole plant |
| 47 | Scrophularia ningpoensis | MeOH | Scrophulariaceae | Whole plant |
| 48 | Catharanthus roseus | MeOH | Apocynaceae | Whole plant above ground |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, H.-W.; Liu, P.-F.; Tsai, W.-L.; Hu, W.-H.; Hu, Y.-C.; Yang, H.-C.; Lin, W.-Y.; Weng, J.-R.; Shu, C.-W. Xanthium strumarium Fruit Extract Inhibits ATG4B and Diminishes the Proliferation and Metastatic Characteristics of Colorectal Cancer Cells. Toxins 2019, 11, 313. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11060313
Chang H-W, Liu P-F, Tsai W-L, Hu W-H, Hu Y-C, Yang H-C, Lin W-Y, Weng J-R, Shu C-W. Xanthium strumarium Fruit Extract Inhibits ATG4B and Diminishes the Proliferation and Metastatic Characteristics of Colorectal Cancer Cells. Toxins. 2019; 11(6):313. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11060313
Chicago/Turabian StyleChang, Hsueh-Wei, Pei-Feng Liu, Wei-Lun Tsai, Wan-Hsiang Hu, Yu-Chang Hu, Hsiu-Chen Yang, Wei-Yu Lin, Jing-Ru Weng, and Chih-Wen Shu. 2019. "Xanthium strumarium Fruit Extract Inhibits ATG4B and Diminishes the Proliferation and Metastatic Characteristics of Colorectal Cancer Cells" Toxins 11, no. 6: 313. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11060313