Regulation of the Staphylococcal Superantigen-Like Protein 1 Gene of Community-Associated Methicillin-Resistant Staphylococcus aureus in Murine Abscesses
Abstract
:1. Introduction
2. Results
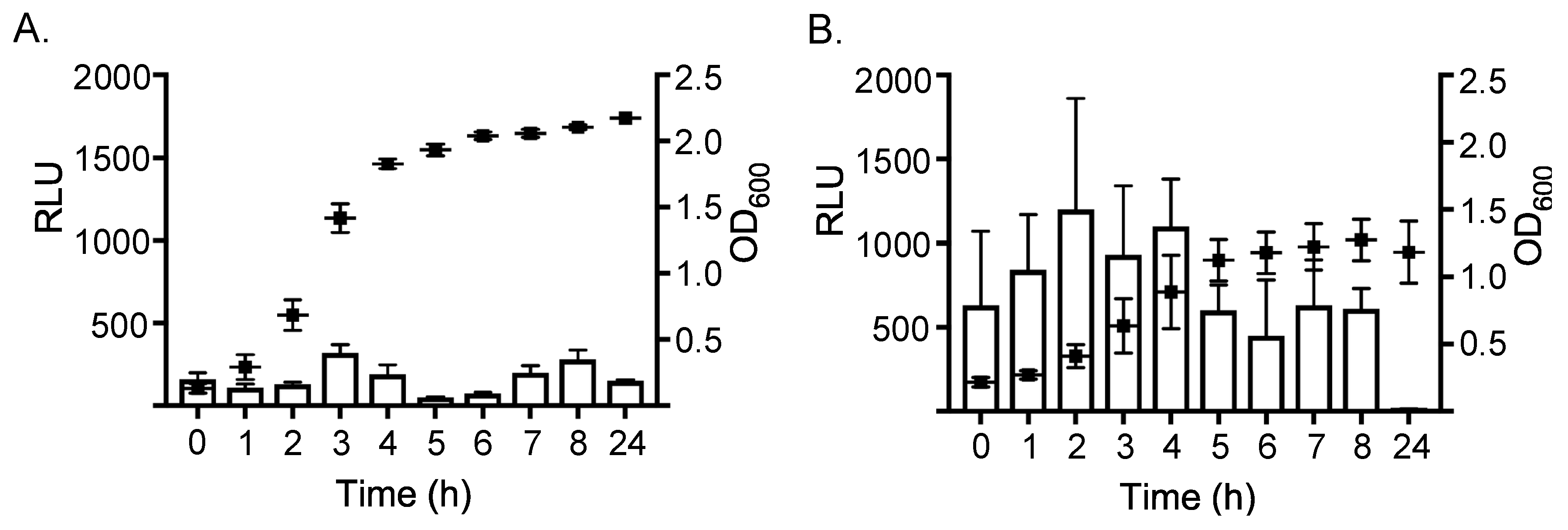
2.1. Expression of ssl1 is Up-Regulated in Nutrient-Poor Conditions
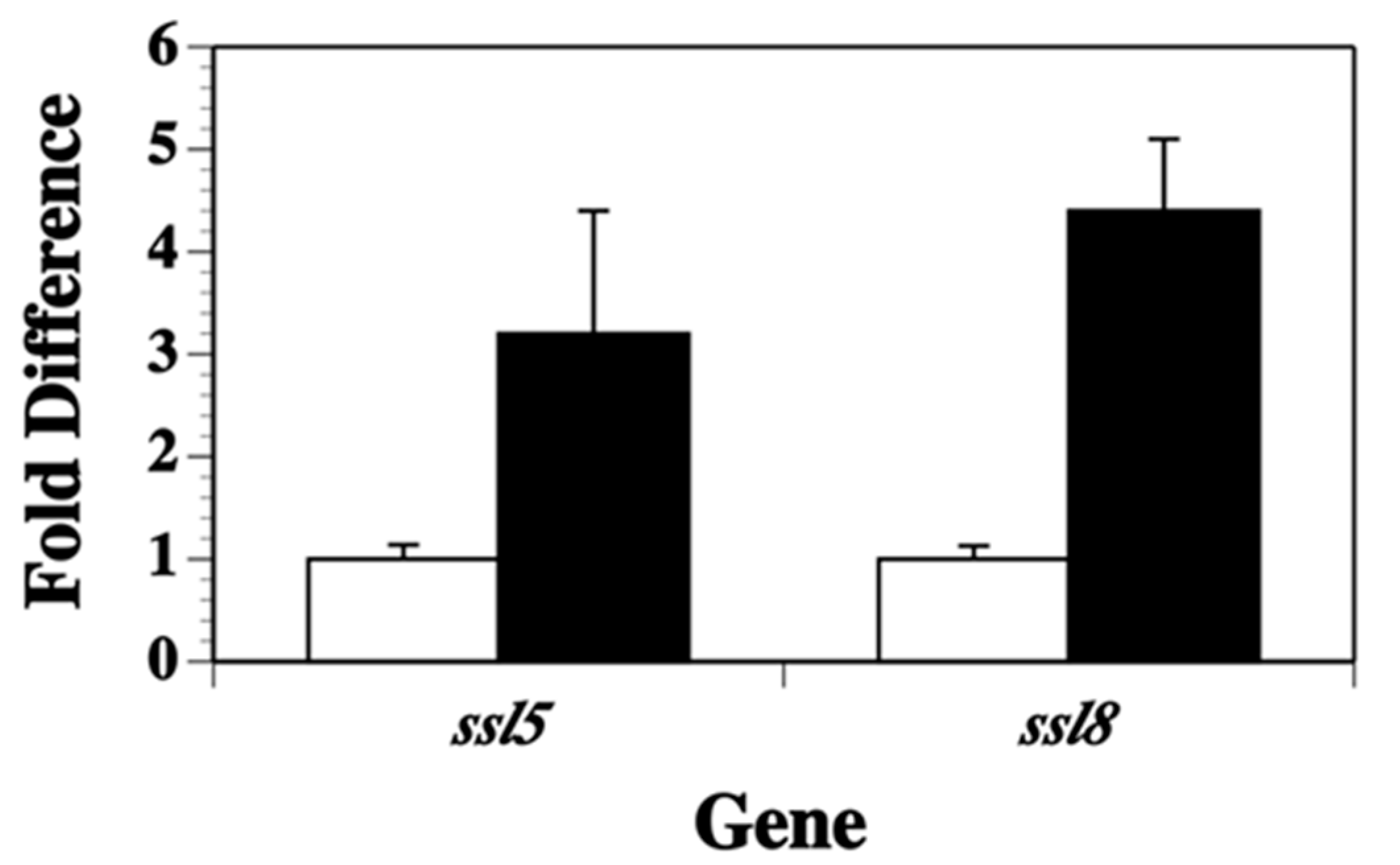
2.2. Transcription of Other ssl Genes Is Up-Regulated in Nutrient Poor Conditions
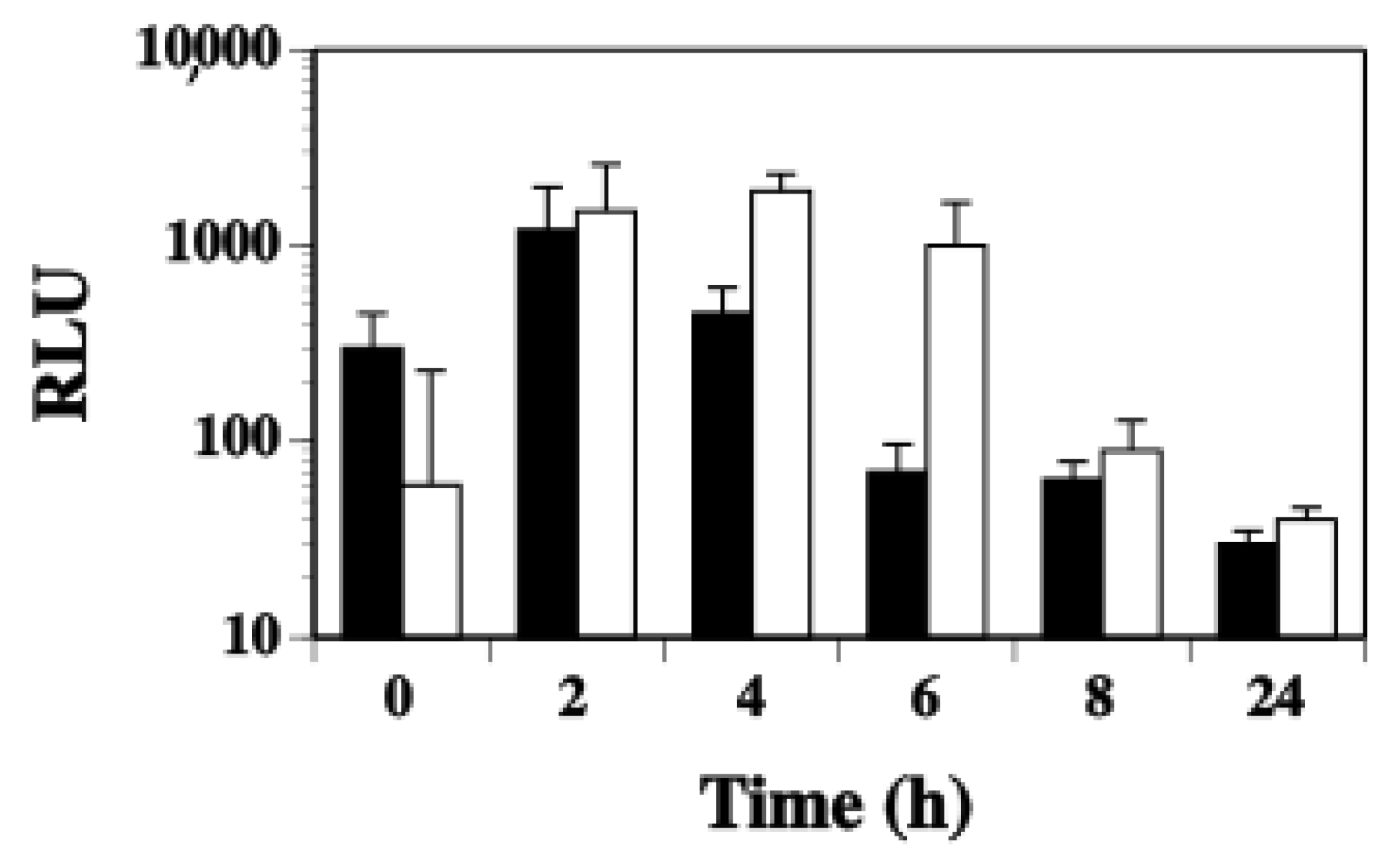
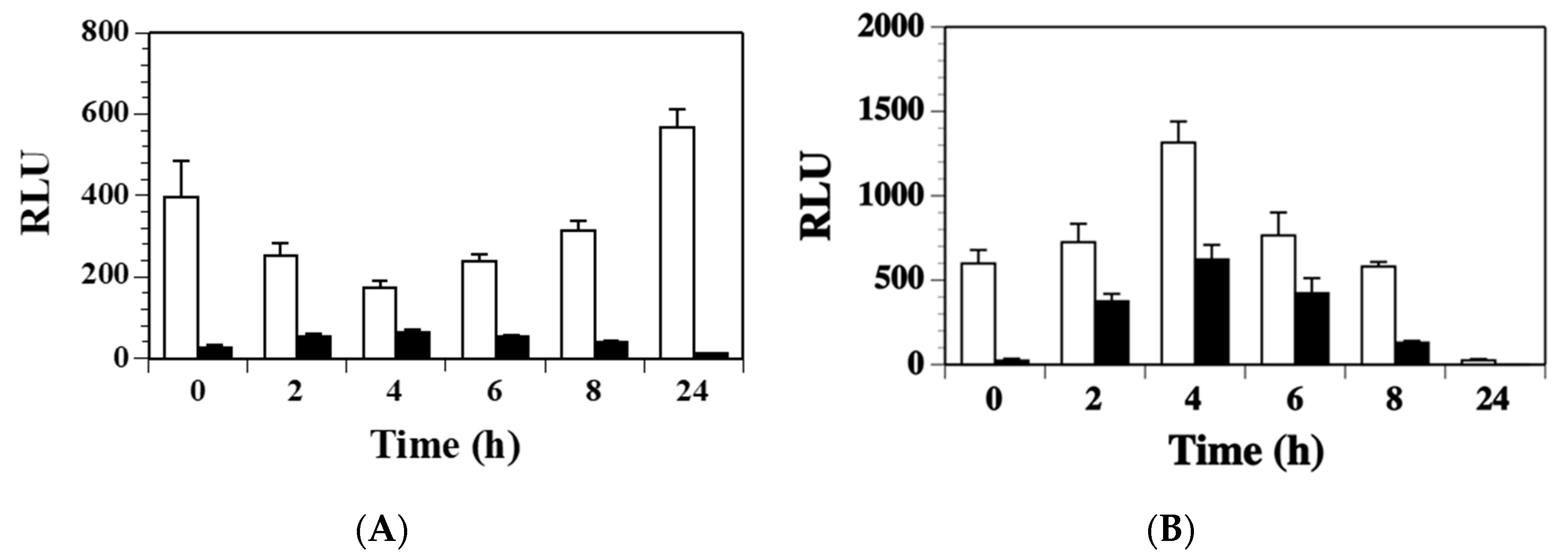
2.3. Agr Has a Minor Contribution to the Modulation of ssl1 Expression in S. aureus
2.4. SaeS Positively Regulates ssl1 Expression in S. aureus
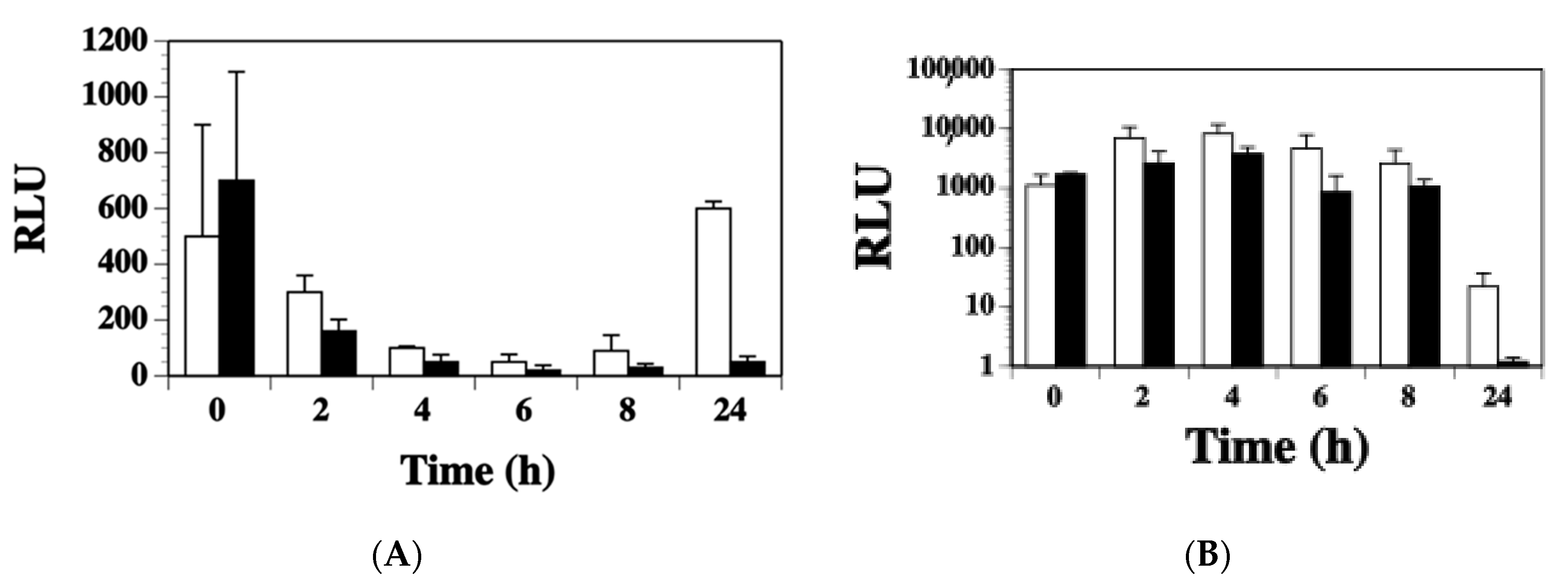
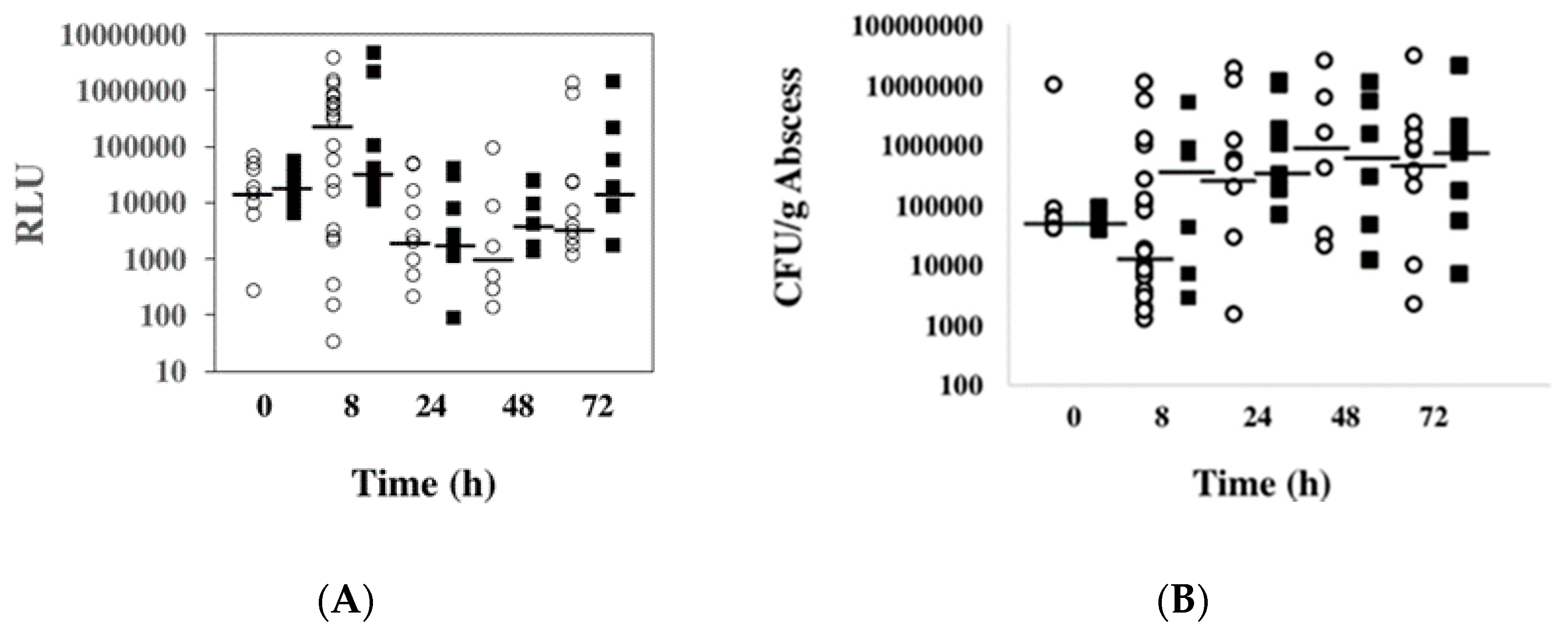
2.5. Expression of ssl1 Occurs Early in S. aureus Found in Murine Abscesses
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains, Plasmids, and Growth Conditions
5.2. Construction of the ssl1::lux Fusion
5.3. Transduction of S. aureus
5.4. In vitro Testing of the ssl1::lux Fusion
5.5. RNA Extraction and Quantitative Reverse Transcribed-Polymerase Chain Reaction (qRT-PCR)
5.6. Murine Abscess Model of Infection
5.7. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Suaya, J.A.; Mera, R.M.; Cassidy, A.; O’Hara, P.; Amrine-Madsen, H.; Burstin, S.; Miller, L.G. Incidence and cost of hospitalizations associated with Staphylococcus aureus skin and soft tissue infections in the United States from 2001 to 2009. BMC Infect. Dis. 2014, 14, 296. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.Y.; Sun, L.; Smith, D.L.; Laxminarayan, R. The changing epidemiology of methicillin-resistant Staphylococcus aureus in the United States: A national observational study. Am. J. Epidemiol. 2013, 1771, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus–Minnesota and North Dakota, 1997-1999. Morbid. Mortal. Wkly. Rep. 1999, 48, 707–710. [Google Scholar]
- Dinges, M.M.; Orwin, P.M.; Schlievert, P.M. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 2000, 13, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Takeuchi, F.; Kuroda, M.; Yuzawa, H.; Aoki, K.; Oguchi, A.; Nagai, Y.; Iwama, N.; Asano, K.; Naimi, T.; et al. Genome and virulence determinant of high virulence community-acquired MRSA. Lancet 2002, 359, 1819–1827. [Google Scholar] [CrossRef]
- Diep, B.A.; Gill, S.R.; Chang, R.G.; Phan, T.H.; Chen, J.H.; Davidson, M.G.; Lin, F.; Lin, J.; Carleton, H.; Mongodin, E.F.; et al. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 2006, 36, 731–739. [Google Scholar] [CrossRef]
- Williams, R.J.; Ward, J.M.; Henderson, B.; Poole, S.; O’Hara, B.P.; Wilson, M.; Nair, S.P. Identification of a novel gene cluster encoding staphylococcal exotoxin-like proteins: Characterization of the prototypic gene and its gene product, SET1. Infect. Immun. 2000, 68, 4407–4415. [Google Scholar] [CrossRef] [PubMed]
- Lina, G.; Bohach, G.A.; Nair, S.P.; Hiramatsu, K.; Jouvin-Marche, E.; Mariuzza, R. Standard nomenclature for the superantigens expressed by Staphylococcus. J. Infect. Dis. 2004, 189, 233–236. [Google Scholar] [CrossRef]
- Holtfreter, S.; Bauer, K.; Thomas, D.; Feig, C.; Lorenz, V.; Roschack, K.; Friebe, K.; Selleng, S.; Lovenich, S.; Greve, T.; et al. egc-encoded superantigens from Staphylococcus aureus are neutralized by human sera much less efficiently than are classical staphylococcal exotoxins or toxic shock syndrome toxin. Infect. Immun. 2004, 72, 4061–4071. [Google Scholar] [CrossRef]
- McCarthy, A.J.; Lindsay, J.A. Staphylococcus aureus innate immune evasion is lineage-specific: A bioinformatics study. Infect. Genet. Evol. 2013, 19, 7–14. [Google Scholar] [CrossRef]
- Langley, R.; Wines, B.; Willoughby, N.; Basu, I.; Proft, T.; Fraser, J.D. The staphylococcal superantigen-like protein 7 binds IgA and complement C5 and inhibits IgA-Fc(alpha)RI binding and serum killing of bacteria. J. Immunol. 2005, 174, 2926–2933. [Google Scholar] [CrossRef] [PubMed]
- Papageorgiou, A.C.; Achaya, K.R. Microbial superantigens: From structure to function. Trends Microbiol. 2000, 8, 369–375. [Google Scholar] [CrossRef]
- Al-Shangiti, A.M.; Naylor, C.E.; Nair, S.P.; Briggs, D.C.; Henderson, B.; Chain, B.M. Structural relationships and cellular tropisms of staphylococcal superantigen-like proteins. Infect. Immun. 2004, 72, 4261–4270. [Google Scholar] [CrossRef] [PubMed]
- Arcus, V.L.; Langley, R.; Proft, R.; Fraser, J.D.; Baker, E.N. The three-dimensional structure of a superantigen-like protein, SET3, from a pathogenicity island of the Staphylococcus aureus genome. J. Biol. Chem. 2002, 277, 32274–32281. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, J.R.; Reid, S.D.; Ruotsalainen, E.; Tripp, T.J.; Liu, M.Y.; Cole, R.; Kuusela, P.; Schlievert, P.M.; Jarvinen, A.; Musser, J.M. Genome diversification in Staphylococcus aureus: Molecular evolution of a highly variable chromosomal region encoding the staphylococcal exotoxin-like family of proteins. Infect. Immun. 2003, 71, 2827–2838. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, N.; Clow, F.; Radcliff, F.J.; Fraser, J.D. Full functional activity of SSL7 requires binding of both complement C5 and IgA. Immunol. Cell Biol. 2013, 91, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Wines, B.D.; Ramsland, P.A.; Trist, H.M.; Gardam, S.; Brink, R.; Fraser, J.D.; Hogarth, P.M. Interaction of human, rat, and mouse immunoglobin A (IgA) with staphylococcal superantigen-like 7 (SSL7) decoy protein and leukocyte IgA receptor. J. Biol. Chem. 2011, 286, 33118–33124. [Google Scholar] [CrossRef] [PubMed]
- Hermans, S.J.; Baker, H.M.; Sequeira, R.P.; Langley, R.J.; Baker, E.N.; Fraser, J.D. Structural and functional properties of staphylococcal superantigen-like protein 4. Infect. Immun. 2012, 80, 4004–4013. [Google Scholar] [CrossRef] [PubMed]
- Itoh, S.; Yamaoka, N.; Kamoshida, G.; Takii, T.; Tsuji, T.; Hayashi, H.; Onozaki, K. Staphylococcal superantigen-like protein 8 (SSL8) binds to tenascin C and inhibits tensacin C-fibronectin interaction and cell motility of keratinocytes. J. Biol. Chem. 2013, 288, 21569–21580. [Google Scholar] [CrossRef]
- Koymans, K.J.; Feitsma, L.J.; Brondijk, T.H.; Aerts, P.C.; Lukkien, E.; Lossl, P.; van Kessel, K.P.; de Haas, C.J.; van Strijp, J.A.; Huizinga, E.G. Structural basis for inhibition of TLR2 by staphylococcal superantigen-like protein 3 (SSL3). Proc. Natl. Acad. Sci. USA 2015, 112, 11018–11023. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, R.; Itoh, S.; Kamoshida, G.; Takii, T.; Fujii, S.; Tsuji, T.; Onozaki, K. Staphylococcal superantigen-like protein 3 binds to the Toll-like receptor 2 extracellular domain and inhibits cytokine production induced by Staphylococcus aureus, cell wall component, or lipopeptides in murine macrophages. Infect. Immun. 2012, 80, 2816–2825. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, M.; Ohta, T.; Uchiyama, I.; Baba, T.; Yuzawa, H.; Kobayashi, I.; Cui, L.; Oguchi, A.; Aoki, K.; Nagai, Y.; et al. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 2001, 357, 1225–1240. [Google Scholar] [CrossRef]
- Al-Shangiti, A.M.; Nair, S.P.; Chain, B.M. The interaction between staphylococcal superantigen-like proteins and dendritic cells. Clin. Exp. Immun. 2005, 140, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Tenover, F.C.; McDougal, L.K.; Goering, R.V.; Killgore, G.; Projan, S.J.; Patel, J.B.; Dunman, P.M. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the Untied States. J. Clin. Microbiol. 2006, 44, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.K.; Karow, M.E.; Brady, J.M.; Stemper, M.E.; Kislow, J.; Moore, N.; Wroblewski, K.; Chyou, P.H.; Warshauer, D.M.; Reed, K.D.; et al. Virulence genes and genotypic associations in nasal carriage, community-associated methicillin-susceptible and methicillin-resistant USA400 Staphylococcus aureus isolates. J. Clin. Microbiol. 2010, 48, 3582–3592. [Google Scholar] [CrossRef]
- Francis, K.P.; Yu, J.; Bellinger-Kawahara, C.; Joh, D.; Hawkinson, M.J.; Xiao, G.; Purchio, T.F.; Caparon, M.G.; Lipitsch, M.; Contag, P.R. Visualizing pneumococcal infections in the lungs of live mice using bioluminescent Streptococcus pneumoniae transformed with a novel gram-positive lux transposon. Infect. Immun. 2001, 69, 3350–3358. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.R. The Staphylococcus as a molecular genetic system. In Molecular Biology of the Staphylococci; Novick, R.P., Ed.; VCH Publishers: New York, NY, USA, 1990; pp. 1–40. [Google Scholar]
- Recsei, P.; Kreiswirth, B.; O’Reilly, M.; Schlievert, P.; Gruss, A.; Novick, R.P. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol. Gen. Genet. 1986, 202, 58–62. [Google Scholar] [CrossRef]
- Yarwood, J.M.; Schlievert, P.M. Quorum sensing in Staphylococcus infections. J. Clin. Investig. 2003, 112, 1620–1625. [Google Scholar] [CrossRef]
- Nygaard, T.K.; Pallister, K.B.; Ruzevich, P.; Griffith, S.; Vunong, C.; Voyich, J.M. SaeR binds a consensus sequence within virulence promoters to advance USA300 pathogenesis. J. Infect. Dis. 2010, 201, 241–254. [Google Scholar] [CrossRef]
- Benson, M.A.; Lilo, S.; Wassaerman, G.A.; Thoendel, M.; Smith, A.; Horswill, A.R.; Fraser, J.; Novick, R.P.; Shopsin, B.; Torres, V.J. Staphylococcus aureus regulates the expression and production of the staphylococcal superantigen-like secreted proteins in a Rot-dependent manner. Mol. Microbiol. 2011, 81, 659–675. [Google Scholar] [CrossRef]
- Sun, F.; Li, C.; Jeong, D.; Sohn, C.; He, C.; Bae, T. In the Staphylococcus aureus two-component sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J. Bacteriol. 2010, 192, 2111–2127. [Google Scholar] [CrossRef] [PubMed]
- Pantrangi, M.; Singh, V.K.; Wolz, C.; Shukla, S.K. Staphylococcal superantigen-like genes, ssl5 and ssl8, are positively regulated by Sae and negatively by Agr in the Newman strain. FEMS Microbiol. Lett. 2010, 308, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Dutta, A.; Bhattacharajee, A.; Basak, A.; Das, A.K. Cloning, expression, crystallization and preliminary X-ray diffraction studies of staphylococcal superantigen-like protein 1 (SSL1). Acta Crystallogr. F Struct. Biol. Commun. 2014, 70, 600–603. [Google Scholar] [CrossRef] [PubMed]
- Koymans, K.J.; Bisschop, A.; Vughs, M.M.; van Kessel, K.P.; de Haas, C.J.; van Strijp, J.A. Staphylococcal superantigen-like protein 1 and 5 (SSL1 & SSL5) limit neutrophil chemotaxis and migration through MMP-inhibition. Int. J. Mol. Sci. 2016, 17, E1072. [Google Scholar] [PubMed]
- Tang, A.; Caballero, A.R.; Bierdeman, M.A.; Marquart, M.E.; Foster, T.J.; Monk, I.R.; O’Callaghan, R.J. Staphylococcus aureus superantigen-like protein SSL1: A toxic protease. Pathogens 2019, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Dunman, P.M.; Murphy, E.; Haney, S.; Palacios, D.; Tucker-Kellogg, G.; Wu, S.; Brown, E.L.; Zagursky, R.J.; Shlaes, D.; Projan, S.J. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 2001, 183, 7341–7353. [Google Scholar] [CrossRef]
- Foster, T.J.; O’Reilly, M.; Phonimdaeng, P.; Cooney, J.; Patel, A.H.; Bramley, A.J. Genetic studies of virulence factors of Staphylococcus aureus. Properties of coagulase and gamma-toxin, alpha-toxin, beta-toxin and protein A in the pathogenesis of S. aureus infections. In Molecular Biology of the Staphylococci; Novick, R.P., Ed.; VCH Publishers: New York, NY, USA, 1990; pp. 403–420. [Google Scholar]
- Laughton, J.M.; Devillard, E.; Heinrichs, D.E.; Reid, G.; McCormick, J.K. Inhibition of expression of a staphylococcal superantigen-like protein by a soluble factor from Lactobacillus reuteri. Microbiology 2006, 152, 1155–1167. [Google Scholar] [CrossRef]
- Tremaine, M.T.; Brockman, D.K.; Betley, M.J. Staphylococcal enterotoxin A gene (sea) expression is not affected by the accessory gene regulator (agr). Infect. Immun. 1993, 61, 356–359. [Google Scholar]
- Zhang, S.; Iandolo, J.J.; Stewart, G.C. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol. Lett. 1998, 168, 227–233. [Google Scholar] [CrossRef]
- Mekalanos, J.J. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 1992, 174, 1–7. [Google Scholar] [CrossRef] [Green Version]
- McNamara, P.J.; Milligan-Monroe, K.C.; Khalili, S.; Proctor, R.A. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 2000, 182, 3197–3203. [Google Scholar] [CrossRef] [PubMed]
- Giraudo, A.T.; Calzolari, A.; Cataldi, A.A.; Bogni, C.; Nagel, R. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 1999, 177, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, C.P.; Boyle-Vavra, S.; Daum, R.S. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS ONE 2010, 5, e15177. [Google Scholar] [CrossRef] [PubMed]
- Voyich, J.M.; Vuong, C.; DeWald, M.; Nygaard, T.K.; Kocianova, S.; Griffith, S.; Jones, J.; Iverson, C.; Sturdevant, D.E.; Braughton, K.R.; et al. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J. Infect. Dis. 2009, 199, 1698–1706. [Google Scholar] [CrossRef] [PubMed]
- Benson, M.A.; Lilo, S.; Nygaard, T.; Voyich, J.M.; Torres, V.J. Rot and SaeRS cooperate to activate expression of the staphylococcal superantigen-like exoproteins. J. Bacteriol. 2012, 194, 4355–4365. [Google Scholar] [CrossRef] [PubMed]
- Besteboer, J.; Poppelier, M.J.J.G.; Ulfman, L.H.; Lenting, P.J.; Denis, C.V.; van Kessel, K.P.M.; van Strijp, J.A.G.; de Haas, C.J.C. Staphylococcal superantigen-like 5 binds PSGL-1 and inhibits P-selectin-mediated neutrophil rolling. Blood 2007, 109, 2936–2943. [Google Scholar]
- Chung, M.C.; Wines, B.D.; Baker, H.; Langley, R.J.; Baker, E.N.; Fraser, J.D. The crystal structure of staphylococcal superantigen-like toxin 11 (SSL11) in complex with sialyl Lewis X reveals the mechanism for cell binding and immune inhibition. Mol. Microbiol. 2007, 66, 1342–1355. [Google Scholar] [CrossRef]
- Swierstra, J.; Debets, S.; de Vogel, C.; Lemmens-den Toom, N.; Verkaik, N.; Ramdani-Bouguessa, N.; Jonkman, M.F.; van Dijl, J.M.; Fahal, A.; van Belkum, A.; et al. IgG4 subclass-specific responses to Staphylococcus aureus antigens shed new light on host-pathogen interaction. Infect. Immun. 2015, 83, 492–501. [Google Scholar] [CrossRef]
- Fey, P.D.; Endres, J.L.; Yajjala, V.K.; Widhelm, T.J.; Boissy, R.J.; Bose, J.L.; Bayles, K.W. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 2013, 4, e00537012. [Google Scholar] [CrossRef]
- Rudin, L.; Sjostrom, J.E.; Lindberg, M.; Philipson, L. Factors affecting competence for transformation in Staphylococcus aureus. J. Bacteriol. 1974, 118, 155–164. [Google Scholar]
- Schwan, W.R.; Wetzel, K.J.; Gomez, T.S.; Stiles, M.A.; Beitlich, B.D.; Grunwald, S. Low-proline environments impair growth, proline transport and in vivo survival of Staphylococcus aureus strain-specific putP mutants. Microbiology 2004, 150, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P.; Ross, H.F.; Projan, S.J.; Kornblum, J.; Kreiswirth, B.; Mohgazeh, S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993, 12, 3967–3975. [Google Scholar] [CrossRef] [PubMed]
- Duthie, E.S.; Lorenz, L.L. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 1952, 6, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Iandolo, J.; Kraemer, G.R. High frequency transformation of Staphylococcus aureus by electroporation. Curr. Microbiol. 1990, 21, 373–376. [Google Scholar]
- Kloos, W.E.; Pattee, P.A. Transduction analysis of the histidine region in Staphylococcus aureus. J. Gen. Microbiol. 1965, 39, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Schwan, W.R.; Polanowski, R.; Dunman, P.M.; Medina-Bielski, S.; Lane, M.; Rott, M.; Lipker, L.; Wescott, A.; Monte, A.; Cook, J.M.; et al. Identification of Staphylococcus aureus cellular pathways affected by the stilbenoid lead drug SK-03-92 using a microarray. Antibiotics 2017, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.S.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Beonton, B.M.; Zhang, J.P.; Pope, C.; Christian, T.; Lee, L.; Winterberg, K.M.; Schmid, M.B.; Buysse, J.M. Large-scale identification of genes required for full virulence of Staphylococcus aureus. J. Bacteriol. 2004, 186, 8478–8489. [Google Scholar] [CrossRef]
| Strain or Plasmid | Relevant Characteristics | Reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | General cloning strain | Invitrogen |
| S. aureus | ||
| MW2 | CA-MRSA USA400 virulent strain | [5] |
| RN4220 | Transformation efficient strain | [27] |
| RN6390 | Virulent strain | [27] |
| RN6911 | Agr inactive, RN6390 with agr::tetM | [54] |
| Newman | Virulent strain | [55] |
| NE1296 | JE2 strain with saeS mutation | [51] |
| WS4108 | RN4220 ssl1::lux | This study |
| WS0501 | RN6390 ssl1::lux | This study |
| WS0604 | RN6390 ssl1::lux, saeS::mariner | This study |
| WS2601 | RN6911 ssl1::lux | This study |
| NS3513 | Newman ssl1::lux | This study |
| Plasmids | ||
| pXen5 | TS a origin, Tn4001, promoterless lux | [26] |
| pXssl | ssl1::lux on pXen5 | This study |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bretl, D.J.; Elfessi, A.; Watkins, H.; Schwan, W.R. Regulation of the Staphylococcal Superantigen-Like Protein 1 Gene of Community-Associated Methicillin-Resistant Staphylococcus aureus in Murine Abscesses. Toxins 2019, 11, 391. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11070391
Bretl DJ, Elfessi A, Watkins H, Schwan WR. Regulation of the Staphylococcal Superantigen-Like Protein 1 Gene of Community-Associated Methicillin-Resistant Staphylococcus aureus in Murine Abscesses. Toxins. 2019; 11(7):391. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11070391
Chicago/Turabian StyleBretl, Daniel J., Abdulaziz Elfessi, Hannah Watkins, and William R. Schwan. 2019. "Regulation of the Staphylococcal Superantigen-Like Protein 1 Gene of Community-Associated Methicillin-Resistant Staphylococcus aureus in Murine Abscesses" Toxins 11, no. 7: 391. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11070391





