Diversity, Cyanotoxin Production, and Bioactivities of Cyanobacteria Isolated from Freshwaters of Greece
Abstract
:1. Introduction
2. Results
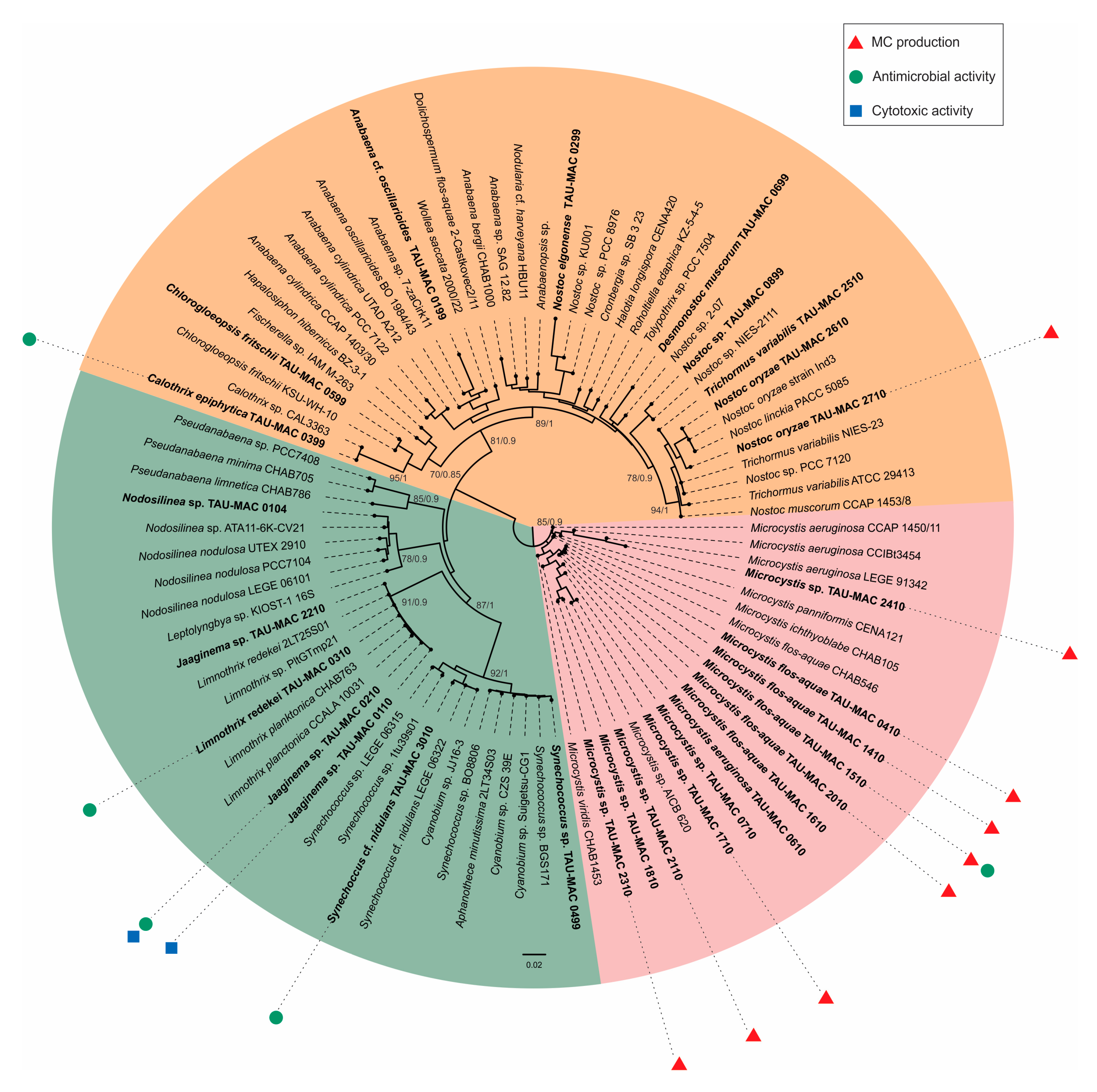
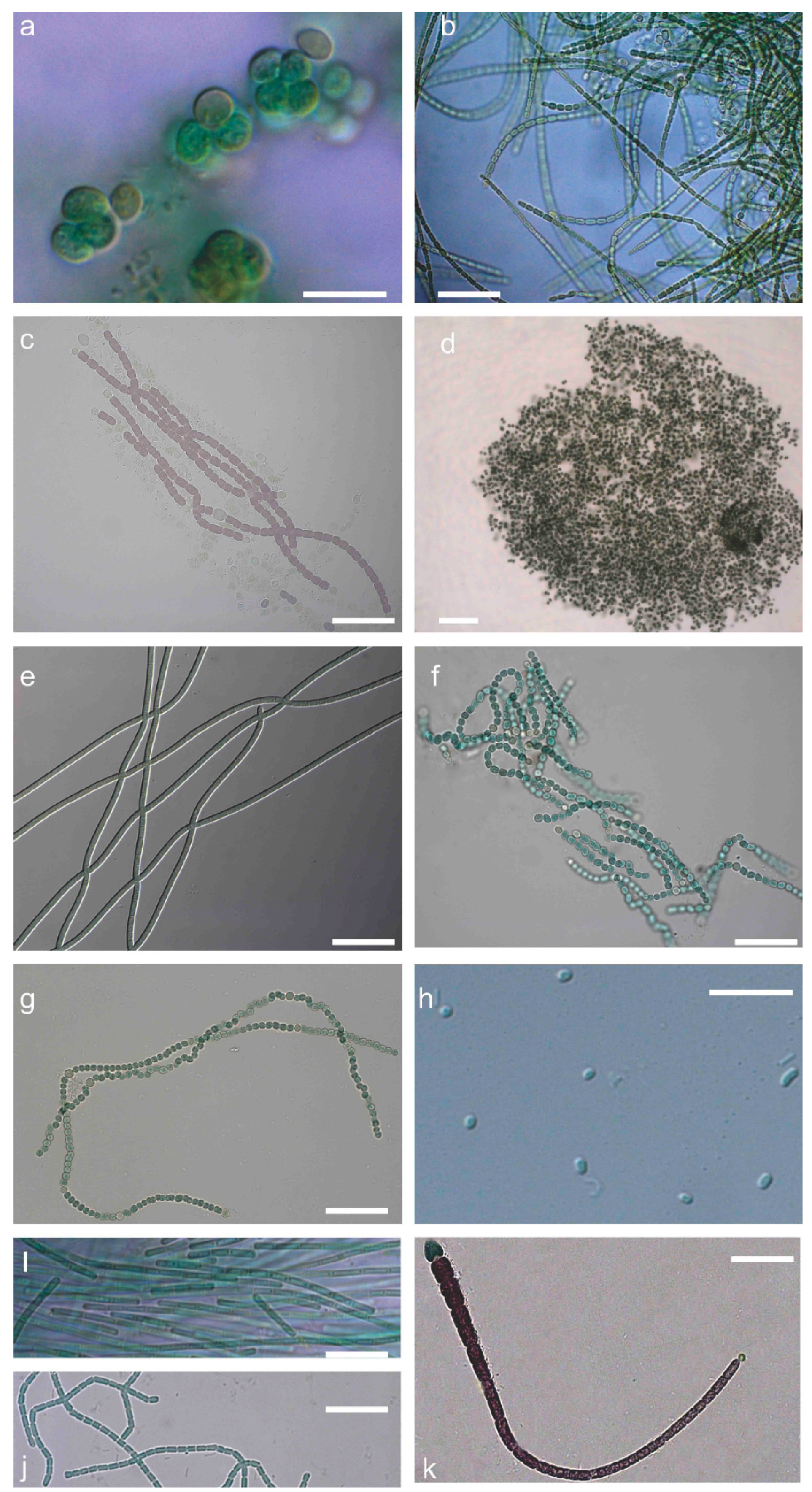
2.1. Polyphasic Taxonomy
2.2. Cyanotoxins
2.3. Antibacterial Activity
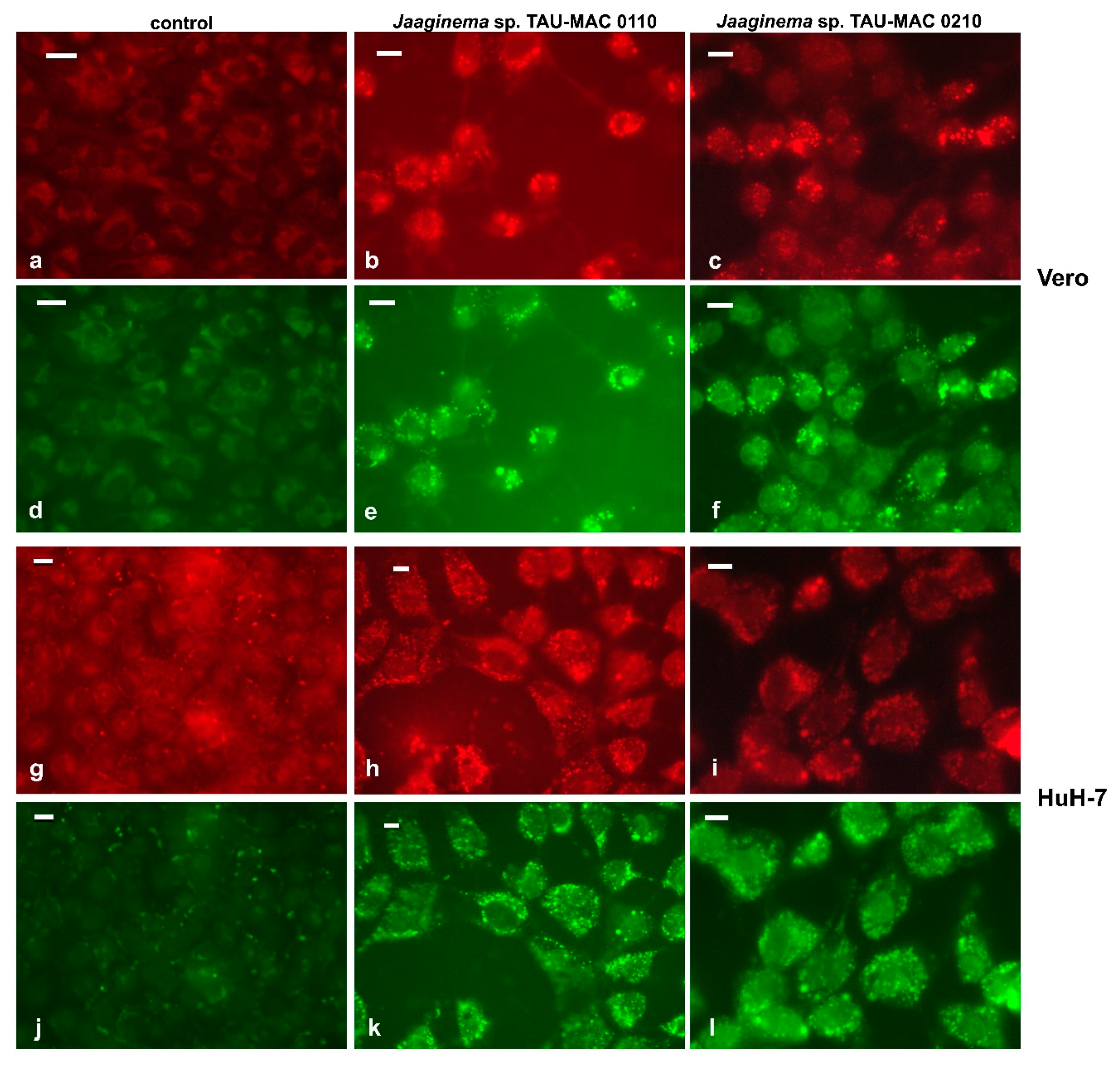
2.4. Effects on Cell Lines
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cyanobacterial Strains and Culture
5.2. Polyphasic Taxonomy
5.3. Cyanotoxin Analysis Using LC–MS/MS
5.4. Extract Preparation for Assays
5.5. Heterotrophic Bacteria and Antibacterial Assays
5.6. Cell Lines and Cytotoxicity Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dittmann, E.; Fewer, D.P.; Neilan, B.A. Cyanobacterial toxins: Biosynthetic routes and evolutionary roots. FEMS Microbiol. Rev. 2013, 37, 23–43. [Google Scholar] [CrossRef]
- Wiegand, C.; Pflugmacher, S. Ecotoxicological effects of selected cyanobacterial secondary metabolites a short review. Toxicol. Appl. Pharmacol. 2005, 203, 201–218. [Google Scholar] [CrossRef]
- Meriluoto, J.; Spoof, L.; Codd, G.A. Handbook of Cyanobacterial Monitoring and Cyanotoxin Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2017; ISBN 1119068681. [Google Scholar]
- Sivonen, K.; Leikoski, N.; Fewer, D.P.; Jokela, J. Cyanobactins-ribosomal cyclic peptides produced by cyanobacteria. Appl. Microbiol. Biotechnol. 2010, 86, 1213–1225. [Google Scholar] [CrossRef]
- Nunnery, J.K.; Mevers, E.; Gerwick, W.H. Biologically active secondary metabolites from marine cyanobacteria. Curr. Opin. Biotechnol. 2010, 21, 787–793. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Parihar, P.; Singh, M.; Bajguz, A.; Kumar, J.; Singh, S.; Singh, V.P.; Prasad, S.M. Uncovering Potential Applications of Cyanobacteria and Algal Metabolites in Biology, Agriculture and Medicine: Current Status and Future Prospects. Front. Microbiol. 2017, 8, 515. [Google Scholar] [CrossRef] [Green Version]
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural Product Biosynthetic Diversity and Comparative Genomics of the Cyanobacteria. Trends Microbiol. 2015, 23, 642–652. [Google Scholar] [CrossRef]
- Burja, A.M.; Banaigs, B.; Abou-Mansour, E.; Grant Burgess, J.; Wright, P.C. Marine cyanobacteria—A prolific source of natural products. Tetrahedron 2001, 57, 9347–9377. [Google Scholar] [CrossRef]
- Welker, M.; Von Döhren, H. Cyanobacterial peptides—Nature’s own combinatorial biosynthesis. FEMS Microbiol. Rev. 2006, 30, 530–563. [Google Scholar] [CrossRef]
- Dixit, R.B.; Suseela, M.R. Cyanobacteria: Potential candidates for drug discovery. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2013, 103, 947–961. [Google Scholar] [CrossRef]
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef]
- Hayes, M.; Bastiaens, L.; Gouveia, L.; Gkelis, S.; Skomedal, H.; Skjanes, K.; Murray, P.; García-Vaquero, M.; Hosoglu, M.I.; Dodd, J. Microalgal Bioactive Compounds Including Protein, Peptides, and Pigments: Applications, Opportunities, and Challenges During Biorefinery Processes. In Novel Proteins for Food, Pharmaceuticals and Agriculture; John Wiley & Sons, Ltd.: Chichester, UK, 2018; pp. 239–255. [Google Scholar]
- D’agostino, G.; Del Campoy, J.; Melladoz, B.; Izquierdo, M.A.; Minarikk, T.; Cirri, L.; Marini, L.; Perez-Gracia, J.L.; Scambia, G. A Multicenter Phase II Study of the Cryptophycin Analog LY355703 in Patients with Platinum-Resistant Ovarian Cancer. Int. J. Gynecol. Cancer 2006, 16, 71–76. [Google Scholar] [CrossRef]
- Liu, L.; Herfindal, L.; Jokela, J.; Shishido, T.K.; Wahlsten, M.; Døskeland, S.O.; Sivonen, K. Cyanobacteria from terrestrial and marine sources contain apoptogens able to overcome chemoresistance in acute myeloid leukemia cells. Mar. Drugs 2014, 12, 2036–2053. [Google Scholar] [CrossRef]
- Deng, R.; Chow, T.-J. Hypolipidemic, antioxidant, and antiinflammatory activities of microalgae Spirulina. Cardiovasc. Ther. 2010, 28, e33–e45. [Google Scholar] [CrossRef]
- Mazur-Marzec, H.; Błaszczyk, A.; Felczykowska, A.; Hohlfeld, N.; Kobos, J.; Toruńska-Sitarz, A.; Devi, P.; Montalvão, S.; D’souza, L.; Tammela, P.; et al. Baltic cyanobacteria—A source of biologically active compounds. Eur. J. Phycol. 2015, 50, 343–360. [Google Scholar] [CrossRef]
- Humisto, A.; Herfindal, L.; Jokela, J.; Karkman, A.; Bjørnstad, R.; Choudhury, R.R.; Sivonen, K. Cyanobacteria as a Source for Novel Anti-Leukemic Compounds. Curr. Pharm. Biotechnol. 2016, 17, 78–91. [Google Scholar] [CrossRef]
- Freitas, S.; Martins, R.; Campos, A.; Azevedo, J.; Osório, H.; Costa, M.; Barros, P.; Vasconcelos, V.; Urbatzka, R. Insights into the potential of picoplanktonic marine cyanobacteria strains for cancer therapies—Cytotoxic mechanisms against the RKO colon cancer cell line. Toxicon 2016, 119, 140–151. [Google Scholar] [CrossRef]
- Engene, N.; Tronholm, A.; Salvador-Reyes, L.A.; Luesch, H.; Paul, V.J. Caldora penicillata gen. nov., comb. nov. (cyanobacteria), a pantropical marine species with biomedical relevance. J. Phycol. 2015, 51, 670–681. [Google Scholar] [CrossRef]
- Costa, M.; Garcia, M.; Costa-Rodrigues, J.; Costa, M.S.; Ribeiro, M.J.; Fernandes, M.H.; Barros, P.; Barreiro, A.; Vasconcelos, V.; Martins, R. Exploring bioactive properties of marine cyanobacteria isolated from the Portuguese coast: High potential as a source of anticancer compounds. Mar. Drugs 2014, 12, 98–114. [Google Scholar] [CrossRef]
- Gkelis, S.; Ourailidis, I.; Panou, M.; Pappas, N. Cyanobacteria of Greece: An annotated checklist. Biodivers. Data J. 2016, 4, e10084. [Google Scholar] [CrossRef]
- Gkelis, S.; Fernández Tussy, P.; Zaoutsos, N. Isolation and preliminary characterization of cyanobacteria strains from freshwaters of Greece. Open Life Sci. 2015, 10, 52–60. [Google Scholar] [CrossRef]
- Gkelis, S.; Lanaras, T.; Sivonen, K. Cyanobacterial toxic and bioactive peptides in freshwater bodies of Greece: Concentrations, occurrence patterns, and implications for human health. Mar. Drugs 2015, 13, 6319–6335. [Google Scholar] [CrossRef]
- Gkelis, S.; Panou, M.; Chronis, I.; Zervou, S.K.; Christophoridis, C.; Manolidi, K.; Ntislidou, C.; Triantis, T.M.; Kaloudis, T.; Hiskia, A.; et al. Monitoring a newly re-born patient: Water quality and cyanotoxin occurrence in a reconstructed shallow Mediterranean lake. Adv. Oceanogr. Limnol. 2017, 8. [Google Scholar] [CrossRef]
- Christophoridis, C.; Zervou, S.K.; Manolidi, K.; Katsiapi, M.; Moustaka-Gouni, M.; Kaloudis, T.; Triantis, T.M.; Hiskia, A. Occurrence and diversity of cyanotoxins in Greek lakes. Sci. Rep. 2018, 8, 17877. [Google Scholar] [CrossRef]
- Sundman, P.; Gugger, M.; Paulin, L.; Vezie, C.; Lyra, C.; Suomalainen, S.; Sivonen, K. Molecular characterization of planktic cyanobacteria of Anabaena, Aphanizomenon, Microcystis and Planktothrix genera. Int. J. Syst. Evol. Microbiol. 2015, 51, 513–526. [Google Scholar]
- Otsuka, S.; Suda, S.; Shibata, S.; Oyaizu, H.; Matsumoto, S.; Watanabe, M.M. A proposal for the unification of five species of the cyanobacterial genus Microcystis Kutzing ex Lemmermann 1907 under the Rules of the Bacteriological Code. Int. J. Syst. Evol. Microbiol. 2001, 51, 873–879. [Google Scholar] [CrossRef]
- Willame, R.; Boutte, C.; Grubisic, S.; Wilmotte, A.; Komárek, J.; Hoffmann, L. Morphological and Molecular Characterization of Planktonic Cyanobacteria From Belgium and Luxembourg 1. J. Phycol. 2006, 42, 1312–1332. [Google Scholar] [CrossRef]
- Valerio, E.; Chambel, L.; Paulino, S.; Faria, N.; Pereira, P.; Tenreiro, R. Molecular identification, typing and traceability of cyanobacteria from freshwater reservoirs. Microbiology 2009, 155, 642–656. [Google Scholar] [CrossRef] [Green Version]
- Neilan, B.A.; Jacobs, D.; Therese, D.D.; Blackall, L.L.; Hawkins, P.R.; Cox, P.T.; Goodman, A.E. rRNA Sequences and Evolutionary Relationships among Toxic and Nontoxic Cyanobacteria of the Genus Microcystis. Int. J. Syst. Bacteriol. 1997, 47, 693–697. [Google Scholar] [CrossRef]
- Kondo, R.; Yoshida, T.; Yuki, Y.; Hiroishi, S. DNA—DNA reassociation among a bloom-forming cyanobacterial genus, Microcystis. Int. J. Syst. Evol. Microbiol. 2000, 50, 767–770. [Google Scholar] [CrossRef]
- Xiao, M.; Li, M.; Reynolds, C.S. Colony formation in the cyanobacterium Microcystis. Biol. Rev. 2018, 93, 1399–1420. [Google Scholar] [CrossRef]
- Perkerson, R.B.; Johansen, J.R.; Kovácik, L.; Brand, J.; Kaštovský, J.; Casamatta, D.A. A unique pseudanabaenalean (cyanobacteria) genus Nodosilinea gen. nov. based on morphological and molecular data. J. Phycol. 2011, 47, 1397–1412. [Google Scholar] [CrossRef] [PubMed]
- Mai, T.; Johansen, J.R.; Pietrasiak, N.; Bohunická, M.; Martin, M.P. Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and Trichocoleaceae fam. nov. and six new genera containing 14 species. Phytotaxa 2018, 365, 1–59. [Google Scholar] [CrossRef] [Green Version]
- Castenholz, R.W.; Wilmotte, A.; Herdman, M.; Rippka, R.; Waterbury, J.B.; Iteman, I.; Hoffmann, L. Phylum BX. Cyanobacteria. In Bergey’s Manual® of Systematic Bacteriology; Springer: New York, NY, USA, 2001; pp. 473–599. [Google Scholar]
- Yongmanitchai, W.; Nopartnaraporn, N.; Suda, S.; Watanabe, M.M.; Liu, Y.; Day, J.G.; Otsuka, S.; Mahakahant, A. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int. J. Syst. Evol. Microbiol. 2002, 52, 1577–1595. [Google Scholar]
- Ishida, T.; Watanabe, M.M.; Sugiyama, J.; Yokota, A. Evidence for polyphyletic origin of the members of the orders of Oscillatoriales and Pleurocapsales as determined by 16S rDNA analysis. FEMS Microbiol. Lett. 2001, 201, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Rajaniemi, P.; Hrouzek, P.; Kaštovská, K.; Willame, R.; Rantala, A.; Hoffmann, L.; Komárek, J.; Sivonen, K. Phylogenetic and morphological evaluation of the genera Anabaena, Aphanizomenon, Trichormus and Nostoc (Nostacales, cyanobacteria). Int. J. Syst. Evol. Microbiol. 2005, 55, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Teneva, I.; Stoyanov, P.; Dimitrova, I. Production of cyanobacterial toxins from two Nostoc species ( Nostocales ) and evaluation of their cytotoxicity in vitro. J. Biosci. Biotechnol. 2012, 1, 33–43. [Google Scholar]
- Bagchi, S.N.; Dubey, N.; Singh, P. Phylogenetically distant clade of Nostoc-like taxa with the description of Aliinostoc gen. nov. and Aliinostoc morphoplasticum sp. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 3329–3338. [Google Scholar] [CrossRef]
- Hindák, F. On Chlorogloeopsis fritschii (Cyanophyta/Cyanobacteria) from thermal springs in Slovakia and from a saline lake in Tunisia. Arch. Hydrobiol. Suppl. Algol. Stud. 2009, 126, 47–64. [Google Scholar] [CrossRef]
- Uher, B.; Kovacik, L.; Katarína, Š.; Jančušová, M.; Jursa, M. The subaerial epilithic species Chlorogloeopsis cf. fritschii (Mitra) Mitra et Pandey (Stigonematales, Cyanobacteria) in National Park Slovak Paradise. Acta Bot. Univ. Comeniensis Bratisl. 2005, 42, 3–6. [Google Scholar]
- Gkelis, S.; Zaoutsos, N. Cyanotoxin occurrence and potentially toxin producing cyanobacteria in freshwaters of Greece: A multi-disciplinary approach. Toxicon 2014, 78, 1–9. [Google Scholar] [CrossRef]
- Chorus, I. Current Approaches to Cyanotoxin Rixk Assessment, Risk Management and Regulations in Different Countries; Federal Environmental Agency (Umweltbundesamt): Berlin, Germany, 2005. [Google Scholar]
- Ríos, V.; Moreno, I.; Prieto, A.I.; Soria-Díaz, M.E.; Frías, J.E.; Cameán, A.M. Comparison of Microcystis aeruginosa (PCC7820 and PCC7806) growth and intracellular microcystins content determined by liquid chromatography-mass spectrometry, enzyme-linked immunosorbent assay anti-Adda and phosphatase bioassay. J. Water Health 2014, 12, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Park, H.D.; Watanabe, M.F.; Harda, K.; Nagai, H.; Suzuki, M.; Watanabe, M.; Hayashi, H. Hepatotoxin (microcystin) and neurotoxin (anatoxin-a) contained in natural blooms and strains of cyanobacteria from Japanese freshwaters. Nat. Toxins 1993, 1, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Cameán, A.; Moreno, I.M.; Ruiz, M.J.; Picó, Y. Determination of microcystins in natural blooms and cyanobacterial strain cultures by matrix solid-phase dispersion and liquid chromatography-mass spectrometry. Anal. Bioanal. Chem. 2004, 380, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Murphy, T.; Guo, J.; Parr, T.; Nalewajko, C. Iron-stimulated growth and microcystin production of Microcystis novacekii UAM 250. Limnologica 2009, 39, 255–259. [Google Scholar] [CrossRef]
- Komárek, J. Heterocytous Genera; Springer: Spektrum, Germany, 2013; ISBN 9783827409324. [Google Scholar]
- Wood, S.A.; Mountfort, D.; Selwood, A.I.; Holland, P.T.; Puddick, J.; Cary, S.C. Widespread distribution and identification of eight novel microcystins in antarctic cyanobacterial mats. Appl. Environ. Microbiol. 2008, 74, 7243–7251. [Google Scholar] [CrossRef] [PubMed]
- Oudra, B.; Dadi-El Andaloussi, M.; Vasconcelos, V.M. Identification and quantification of microcystins from a Nostoc muscorum bloom occurring in Oukaïmeden River (High-Atlas mountains of Marrakech, Morocco). Environ. Monit. Assess. 2009, 149, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Genuário, D.B.; Silva-Stenico, M.E.; Welker, M.; Beraldo Moraes, L.A.; Fiore, M.F. Characterization of a microcystin and detection of microcystin synthetase genes from a Brazilian isolate of Nostoc. Toxicon 2010, 55, 846–854. [Google Scholar] [CrossRef]
- Yanni, Y.G.; Osman, Z.H. Contributions of algalization to rice growth, yield, N attributes and incidence of infestation with the blast fungus Pyricularia oryzae under different fungicidal treatments. World J. Microbiol. Biotechnol. 1990, 6, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Han, F.X.; Wang, F.; Zhang, H.; Shi, Z. Accumulation and phytotoxicity of microcystin-LR in rice (Oryza sativa). Ecotoxicol. Environ. Saf. 2012, 76, 193–199. [Google Scholar] [CrossRef]
- Kreitlow, S.; Mundt, S.; Lindequist, U. Cyanobacteria—A potential source of new biologically active substances. J. Biotechnol. 1999, 70, 61–63. [Google Scholar] [CrossRef]
- Martins, R.F.; Ramos, M.F.; Herfindal, L.; Sousa, J.A.; Skaerven, K.; Vasconcelos, V.M. Antimicrobial and cytotoxic assessment of marine cyanobacteria—Synechocystis and Synechococcus. Mar. Drugs 2008, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.; Al-Nazawi, M.; Alderson, G. Permeabilising effects of sub-inhibitory concentrations of microcystin on the growth of Escherichia coli. FEMS Microbiol. Lett. 2004, 230, 167–170. [Google Scholar] [CrossRef]
- Issa, A.A. Antibiotic production by the cyanobacteria Oscillatoria angustissima and Calothrix parietina. Environ. Toxicol. Pharmacol. 1999, 8, 33–37. [Google Scholar] [CrossRef]
- Pushparaj, B.; Pelosi, E.; Jüttner, F. Toxicological analysis of the marine cyanobacterium Nodularia harveyana. J. Appl. Phycol. 1998, 10, 527–530. [Google Scholar] [CrossRef]
- Fastner, J.; Heinze, R.; Humpage, A.; Mischke, U.; Eaglesham, G.; Chorus, I. Cylindrospermopsin occurrence in two German lakes and preliminary assessment of toxicity and toxin production of Cylindrospermopsis raciborskii (Cyanobacteria) isolates. Toxicon 2003, 42, 313–321. [Google Scholar] [CrossRef]
- Jerez-Martel, I.; García-Poza, S.; Rodríguez-Martel, G.; Rico, M.; Afonso-Olivares, C.; Gómez-Pinchetti, J.L. Phenolic profile and antioxidant activity of crude extracts from microalgae and cyanobacteria strains. J. Food Qual. 2017, 2017, 2924508. [Google Scholar] [CrossRef]
- Gardel, M.L.; Nakamura, F.; Hartwig, J.H.; Crocker, J.C.; Stossel, T.P.; Weitz, D.A. Prestressed F-actin networks cross-linked by hinged filamins replicate mechanical properties of cells. Proc. Natl. Acad. Sci. USA 2006, 103, 1762–1767. [Google Scholar] [CrossRef] [Green Version]
- Carter, C.A.; Hamm, J.T. Multiplexed quantitative high content screening reveals that cigarette smoke condensate induces changes in cell structure and function through alterations in cell signaling pathways in human bronchial cells. Toxicology 2009, 261, 89–102. [Google Scholar] [CrossRef]
- Carter, C.A.; Madden, V.J. A Newly Characterized Human Endometrial Adenocarcinoma Cell Line (CAC-1) Differentiates in Response to Retinoic Acid Treatment. Exp. Mol. Pathol. 2000, 69, 175–191. [Google Scholar] [CrossRef]
- Frisch, S.M.; Francis, H. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 1994, 124, 619–626. [Google Scholar] [CrossRef] [Green Version]
- Herfindal, L.; Oftedal, L.; Selheim, F.; Wahlsten, M.; Sivonen, K.; Døskeland, S.O. A high proportion of Baltic Sea benthic cyanobacterial isolates contain apoptogens able to induce rapid death of isolated rat hepatocytes. Toxicon 2005, 46, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Hrouzek, P.; Kapuścik, A.; Vacek, J.; Voráčová, K.; Paichlová, J.; Kosina, P.; Voloshko, L.; Ventura, S.; Kopecký, J. Cytotoxicity evaluation of large cyanobacterial strain set using selected human and murine in vitro cell models. Ecotoxicol. Environ. Saf. 2016, 124, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Ge, X.; Gao, Y.; Herrler, G.; Ren, Y.; Ren, X.; Li, G. Porcine epidemic diarrhea virus inhibits dsRNA-induced interferon-β production in porcine intestinal epithelial cells by blockade of the RIG-I-mediated pathway. Virol. J. 2015, 12, 127. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Prins, R.M.; Dang, J.; Kuga, D.; Iwanami, A.; Soto, H.; Lin, K.Y.; Huang, T.T.; Akhavan, D.; Hock, M.B.; et al. EGFR Signaling Through an Akt-SREBP-1-Dependent, Rapamycin-Resistant Pathway Sensitizes Glioblastomas to Antilipogenic Therapy. Sci. Signal. 2009, 2, ra82. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, T.; Parton, R.G. Not Just Fat: The Structure and Function of the Lipid Droplet. Cold Spring Harb. Perspect. Biol. 2011, 3, a004838. [Google Scholar] [CrossRef] [PubMed]
- Przybytkowski, E.; Behrendt, M.; Dubois, D.; Maysinger, D. Nanoparticles can induce changes in the intracellular metabolism of lipids without compromising cellular viability. FEBS J. 2009, 276, 6204–6217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vickers, A.E.M. Characterization of hepatic mitochondrial injury induced by fatty acid oxidation inhibitors. Toxicol. Pathol. 2009, 37, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Ramos, D.F.; Matthiensen, A.; Colvara, W.; de Votto, A.P.S.; Trindade, G.S.; da Silva, P.E.A.; Yunes, J.S. Antimycobacterial and cytotoxicity activity of microcystins. J. Venom. Anim. Toxins Incl. Trop. Dis. 2015, 21, 9. [Google Scholar] [CrossRef]
- Botha, N.; Gehringer, M.M.; Downing, T.G.; Van De Venter, M.; Shephard, E.G. The role of microcystin-LR in the induction of apoptosis and oxidative stress in CaCo2 cells. Toxicon 2004, 43, 85–92. [Google Scholar] [CrossRef]
- Silva-Stenico, M.E.; Silva, C.S.P.; Lorenzi, A.S.; Shishido, T.K.; Etchegaray, A.; Lira, S.P.; Moraes, L.A.B.; Fiore, M.F. Non-ribosomal peptides produced by Brazilian cyanobacterial isolates with antimicrobial activity. Microbiol. Res. 2011, 166, 161–175. [Google Scholar] [CrossRef]
- Asthana, R.K.; Srivastava, A.; Kayastha, A.M.; Nath, G.; Singh, S.P. Antibacterial potential of γ-linolenic acid from Fischerella sp. colonizing Neem tree bark. World J. Microbiol. Biotechnol. 2006, 22, 443–448. [Google Scholar] [CrossRef]
- Nagarajan, M.; Maruthanayagam, V.; Sundararaman, M. A review of pharmacological and toxicological potentials of marine cyanobacterial metabolites. J. Appl. Toxicol. 2012, 32, 153–185. [Google Scholar] [CrossRef] [PubMed]
- Welker, M.; Brunke, M.; Preussel, K.; Lippert, I.; von Döhren, H. Diversity and distribution of Microcystis (cyanobacteria) oligopeptide chemotypes from natural communities studies by single-colony mass spectrometry. Microbiology 2004, 150, 1785–1796. [Google Scholar] [CrossRef] [PubMed]
- Rzymski, P.; Brygider, A.; Kokociński, M. On the occurrence and toxicity of Cylindrospermopsis raciborskii in Poland. Limnol. Rev. 2017, 17, 23–29. [Google Scholar] [CrossRef]
- Regueiras, A.; Pereira, S.; Sofia Costa, M.; Vasconcelos, V. Differential Toxicity of Cyanobacteria Isolated from Marine Sponges towards Echinoderms and Crustaceans. Toxins 2018, 10, 297. [Google Scholar] [CrossRef] [PubMed]
- Le Manach, S.; Duval, C.; Marie, A.; Djediat, C.; Catherine, A.; Edery, M.; Bernard, C.; Marie, B. Global Metabolomic Characterizations of Microcystis spp. Highlights Clonal Diversity in Natural Bloom-Forming Populations and Expands Metabolite Structural Diversity. Front. Microbiol. 2019, 10, 791. [Google Scholar] [CrossRef] [PubMed]
- Kaebernick, M.; Rohrlack, T.; Christoffersen, K.; Neilan, B.A. A spontaneous mutant of microcystin biosynthesis: Genetic characterization and effect on Daphnia. Environ. Microbiol. 2001, 3, 669–679. [Google Scholar] [CrossRef]
- Sogge, H.; Rohrlack, T.; Rounge, T.B.; Sønstebø, J.H.; Tooming-Klunderud, A.; Kristensen, T.; Jakobsen, K.S. Gene Flow, Recombination, and Selection in Cyanobacteria: Population Structure of Geographically Related Planktothrix Freshwater Strains. Appl. Environ. Microbiol. 2013, 79, 508–515. [Google Scholar] [CrossRef]
- Wilson, A.E.; Sarnelle, O.; Neilan, B.A.; Salmon, T.P.; Gehringer, M.M.; Hay, M.E. Genetic variation of the bloom-forming Cyanobacterium Microcystis aeruginosa within and among lakes: Implications for harmful algal blooms. Appl. Environ. Microbiol. 2005, 71, 6126–6133. [Google Scholar] [CrossRef]
- Turner, A.D.; Dhanji-Rapkova, M.; O’Neill, A.; Coates, L.; Lewis, A.; Lewis, K. Analysis of Microcystins in Cyanobacterial Blooms from Freshwater Bodies in England. Toxins 2018, 10, 39. [Google Scholar] [CrossRef]
- Gkelis, S.; Panou, M. Capturing biodiversity: Linking a cyanobacteria culture collection to the scratchpads virtual research environment enhances biodiversity knowledge. Biodivers. Data J. 2016, 4, e7965-e1. [Google Scholar] [CrossRef] [PubMed]
- Rippka, R. Isolation and purification of cyanobacteria. Methods Enzymol. 1988, 167, 3–27. [Google Scholar] [PubMed]
- Komárek, J.; Anagnostidis, K. Süßwasserflora von Mitteleuropa, Bd. 19/2: Cyanoprokaryota: Bd. 2/Part 2: Oscillatoriales; Springer: Spektrum, Germany, 2007; ISBN 9783827419149. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swofford, D.L. Phylogenetic Analysis Using Parsimony. Options 2002, 42, 294–307. [Google Scholar]
- Rambaut, A. FigTree. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 7 July 2018).
- Zervou, S.K.; Christophoridis, C.; Kaloudis, T.; Triantis, T.M.; Hiskia, A. New SPE-LC-MS/MS method for simultaneous determination of multi-class cyanobacterial and algal toxins. J. Hazard. Mater. 2017, 323, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Atlas, R.M. Handbook of Media for Environmental Microbiology; Taylor & Francis: Milton Park, UK, 2005; ISBN 9780849335600. [Google Scholar]
- Kullak-Ublick, G.A.; Beuers, U.; Paumgartner, G. Molecular and functional characterization of bile acid transport in human hepatoblastoma HepG2 cells. Hepatology 1996, 23, 1053–1060. [Google Scholar] [CrossRef]
- Menezes, C.; Valerio, E.; Dias, E. The Kidney Vero-E6 Cell Line: A Suitable Model to Study the Toxicity of Microcystins. In New Insights into Toxicity and Drug Testing; InTech: London, UK, 2013. [Google Scholar] [Green Version]
- Leisching, G.; Loos, B.; Botha, M.; Engelbrecht, A.-M. Bcl-2 confers survival in cisplatin treated cervical cancer cells: Circumventing cisplatin dose-dependent toxicity and resistance. J. Transl. Med. 2015, 13, 328. [Google Scholar] [CrossRef]
- Riss, T.L.; Moravec, R.A.; Niles, A.L.; Duellman, S.; Benink, H.A.; Worzella, T.J.; Minor, L. Cell Viability Assays; Eli Lilly & Company: Indianapolis, IN, USA; National Center for Advancing Translational Sciences: Bethesda, MA, USA, 2004.
- Panteris, E. Cortical actin filaments at the division site of mitotic plant cells: A reconsideration of the “actin-depleted zone”. New Phytol. 2008, 179, 334–341. [Google Scholar] [CrossRef]



| Strain | Toxin Concentration (μg∙g−1 Dry Weight) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [D-Asp3] MC-RR | MC-RR | MC-YR | MC-HtyR | [D-Asp3] MC-LR | MC-LR | MC-HilR | MC-WR | MC-LA | MC-LY | MC-LW | MC-LF | Total | |
| Microcystis aeruginosa TAU-MAC 0610 | -* | - | - | - | - | - | - | - | - | - | - | - | |
| Microcystis flos-aquae TAU-MAC 0410 | - | - | 1029.6 | - | 366.4 | 728.4 | 48.3 | - | <LOQ | <LOQ | 2.3 | - | 2175.0 |
| Microcystis flos-aquae TAU-MAC 1410 | - | - | 1379.6 | - | 227.4 | 809.2 | 50.0 | - | <LOQ | <LOQ | 2.6 | - | 2468.8 |
| Microcystis flos-aquae TAU-MAC 1510 | - | - | 704.7 | - | 293.8 | 489.0 | 32.9 | - | <LOQ | <LOQ | <LOQ | - | 1520.4 |
| Microcystis flos-aquae TAU-MAC 1610 | - | - | - | - | - | - | - | - | - | - | |||
| Microcystis flos-aquae TAU-MAC 2010 | - | - | 860.9 | - | 241.6 | 535.5 | 33.4 | - | <LOQ | tr. | 1.9 | - | 1673.3 |
| Microcystis viridis TAU-MAC 1810 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Microcystis sp. TAU-MAC 0710 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Microcystis sp. TAU-MAC 1710 | - | - | 434.6 | - | 55.90 | 317.7 | 22.2 | - | <LOQ | <LOQ | 2.6 | - | 833.0 |
| Microcystis sp. TAU-MAC 2110 | - | - | 1983.7 | - | 347.6 | 1257.1 | 79.7 | - | 1.2 | 2.9 | 8.7 | - | 3680.9 |
| Microcystis sp. TAU-MAC 2310 | 11.6 | 360.6 | 445.7 | - | 10.7 | 269.8 | 54.0 | 54.6 | - | - | - | 1207.0 | |
| Microcystis sp. TAU-MAC 2410 | - | - | 1533.5 | - | 430.3 | 1105.8 | 64.3 | - | <LOQ | 1.8 | 4.3 | <LOQ | 3140.0 |
| Synechococcus sp. TAU-MAC 0499 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Synechococcus cf. nidulans TAU-MAC 3010 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Jaaginema sp. TAU-MAC 0110 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Jaaginema sp. TAU-MAC 0210 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Jaaginema sp. TAU-MAC 2210 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Limnothrix redekei TAU-MAC 0310 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Nodosilinea sp. TAU-MAC 0104 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Anabaena cf. oscillarioides TAU-MAC 0199 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Chlorogloeopsis fritschii TAU-MAC 0599 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Desmonostoc muscorum TAU-MAC 0699 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Nostoc elgonense TAU-MAC 0299 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Nostoc oryzae TAU-MAC 2610 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Nostoc oryzae TAU-MAC 2710 | - | - | 1.6 | - | <LOQ | <LOQ | - | - | - | - | - | - | 1.6 |
| Nostoc sp. TAU-MAC 0799 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Nostoc sp. TAU-MAC 0899 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Trichormus variabilis TAU-MAC 2510 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Calothrix epiphytica TAU-MAC 0399 | - | - | - | - | - | - | - | - | - | - | - | - | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gkelis, S.; Panou, M.; Konstantinou, D.; Apostolidis, P.; Kasampali, A.; Papadimitriou, S.; Kati, D.; Di Lorenzo, G.M.; Ioakeim, S.; Zervou, S.-K.; et al. Diversity, Cyanotoxin Production, and Bioactivities of Cyanobacteria Isolated from Freshwaters of Greece. Toxins 2019, 11, 436. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11080436
Gkelis S, Panou M, Konstantinou D, Apostolidis P, Kasampali A, Papadimitriou S, Kati D, Di Lorenzo GM, Ioakeim S, Zervou S-K, et al. Diversity, Cyanotoxin Production, and Bioactivities of Cyanobacteria Isolated from Freshwaters of Greece. Toxins. 2019; 11(8):436. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11080436
Chicago/Turabian StyleGkelis, Spyros, Manthos Panou, Despoina Konstantinou, Panagiotis Apostolidis, Antonia Kasampali, Sofia Papadimitriou, Dominiki Kati, Giorgia Maria Di Lorenzo, Stamatia Ioakeim, Sevasti-Kiriaki Zervou, and et al. 2019. "Diversity, Cyanotoxin Production, and Bioactivities of Cyanobacteria Isolated from Freshwaters of Greece" Toxins 11, no. 8: 436. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11080436







