Bottom-Up Proteomic Analysis of Polypeptide Venom Components of the Giant Ant Dinoponera Quadriceps
Abstract
:1. Introduction
2. Results
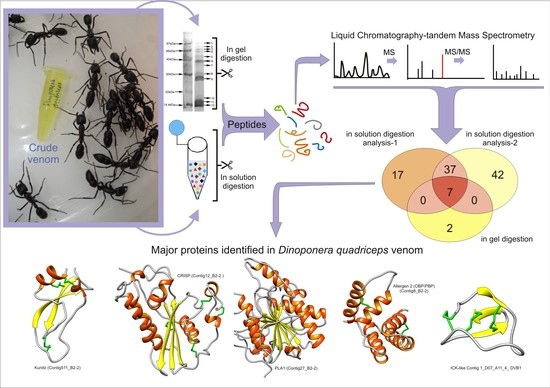
2.1. Bottom-Up Proteomics of Dinoponera quadriceps Venom
2.2. Comparison of Identified Venom-Derived Polypeptides
2.2.1. Venom Dipeptidyl Peptidase-4 (vDPP-4)
2.2.2. Glucose Dehydrogenase [FAD, Quinone]
2.2.3. Yellow Royal Jelly Protein Domain
2.2.4. Phospholipases A1 (PLA1)
2.2.5. Hyaluronidase
2.2.6. Major Venom Allergen 3, Cysteine-Rich Venom Protein Superfamily; Cysteine-Rich Secretary Protein (CRISP) Family
2.2.7. Major Ant Venom Allergen 2/4-Like (Odorant/Pheromone-Binding Protein-Like)
2.2.8. Dinoponera quadriceps Bovine Pancreatic Trypsin Inhibitor (BPTI)/Kunitz-Like Serine Protease Inhibitor
2.2.9. Pilosulin- and Ponericin-Like Peptides
2.2.10. Inhibitor Cysteine Knot (ICK)-Like Venom-Peptides
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Ant Sampling and Venom Extraction
5.2. Proteomic Analysis
5.2.1. Separation of Venom Protein by Denaturing Polyacrylamide Gel Electrophoresis (PAGE)
5.2.2. In Gel Digestion and Mass Spectrometry Analyze
5.2.3. In-Solution Digestion and Mass Spectrometry Analysis
Analysis #1
Analysis #2
5.3. Data Processing and Data Analysis
5.3.1. In-Gel Digestion
5.3.2. In-Solution Digestion
- (a)
- only one protein for each group (each group containing the proteins identified by a common set of peptides was maintained—Peaks software classification);
- (b)
- If one group contained more than one identified protein, the first protein was maintained and the other protein hits were considered redundant and, thus, were removed;
- (c)
- Despite the item b, additional 9 groups were left aside because they contained redundant sequences (three groups from the “in-solution analysis (i)” data and 6 groups from the “in-solution analysis (ii)” data);
- (d)
- both in-solution datasets and highlighted proteins containing the same contig names were compared;
5.4. Structural Models of Selected Dinoponera Quadriceps Venom Toxin
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BPTI | Bovine Pancreatic Trypsin Inhibitor |
| CRISP | Cysteine-rich secretory proteins |
| ICK | Inhibitor Cysteine Knott (Knottin) |
| LC-ESI-MS | Liquid chromatography-electrospray ionization mass spectrometry |
| MRJP | Major royal jelly protein |
| PDB | Protein Data Bank |
| PLA1 | Phospholipase 1 |
| PLA2 | Phospholipase 2 |
| SCP | SCP-like extracellular protein domain |
| CAP | Cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| Sol i 1 | Venom Allergen 1, phospholipase A1, from the fire ant Solenopsis invicta |
| Sol i 2 | Venom Allergen 2 from the fire ant S. invicta |
| Sol i 3 | Venom Allergen 3, SCP/CRISP-like protein, from the fire ant S. invicta |
| Sol i 4 | Venom Allergen 4 from the fire ant S. invicta |
References
- Wilson, R.; Gullan, P.J.; Cranston, P.S. The Insects: An Outline of Entomology, 4th ed.; Wiley: Hoboken, NJ, USA, 2010; Volume 14. [Google Scholar]
- Fernández, F.; Ospina, M. Sinopsis de las Hormigas de la Región Neotropical; Instituto de Investigación de Recursos Biológicos Alexander Von Humbolt: Bogotá, Colombia, 2003. [Google Scholar]
- Touchard, A.; Aili, S.R.; Fox, E.G.; Escoubas, P.; Orivel, J.; Nicholson, G.M.; Dejean, A. The Biochemical Toxin Arsenal from Ant Venoms. Toxins (Basel) 2016, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Schoeters, E.; Billen, J. Morphology and ultrastructure of the convoluted gland in the ant Dinoponera australis (Hymenoptera: Formicidae). Int. J. Insect Morphol. Embryol. 1995, 24, 323–332. [Google Scholar] [CrossRef]
- Robinson, S.D.; Mueller, A.; Clayton, D.; Starobova, H.; Hamilton, B.R.; Payne, R.J.; Vetter, I.; King, G.F.; Undheim, E.A.B. A comprehensive portrait of the venom of the giant red bull ant, Myrmecia gulosa, reveals a hyperdiverse hymenopteran toxin gene family. Sci. Adv. 2018, 4, eaau4640. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.R.; Rikli, H.G.; Schmidt, J.O.; Evans, M.S. A reexamination of poneratoxin from the venom of the bullet ant Paraponera clavata. Peptides 2017, 98, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos Pinto, J.R.; Fox, E.G.; Saidemberg, D.M.; Santos, L.D.; da Silva Menegasso, A.R.; Costa-Manso, E.; Machado, E.A.; Bueno, O.C.; Palma, M.S. Proteomic view of the venom from the fire ant Solenopsis invicta Buren. J. Proteome Res. 2012, 11, 4643–4653. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, S.A.; Haghipour-Peasley, J.; Hoffman, D.R.; Deslippe, R.J. Identification, expression, and immuno-reactivity of Sol i 2 & Sol i 4 venom proteins of queen red imported fire ants, Solenopsis invicta Buren (Hymenoptera: Formicidae). Toxicon 2012, 60, 752–759. [Google Scholar] [CrossRef]
- Aili, S.R.; Touchard, A.; Escoubas, P.; Padula, M.P.; Orivel, J.; Dejean, A.; Nicholson, G.M. Diversity of peptide toxins from stinging ant venoms. Toxicon 2014, 92, 166–178. [Google Scholar] [CrossRef]
- Pluzhnikov, K.A.; Kozlov, S.A.; Vassilevski, A.A.; Vorontsova, O.V.; Feofanov, A.V.; Grishin, E.V. Linear antimicrobial peptides from Ectatomma quadridens ant venom. Biochimie 2014, 107 Pt B, 211–215. [Google Scholar] [CrossRef]
- Kikuchi, K.; Sugiura, M.; Kimura, T. High Proteolytic Resistance of Spider-Derived Inhibitor Cystine Knots. Int. J. Pept. 2015, 2015, 8. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Takashima, H.; Tamaoki, H.; Kyogoku, Y.; Lambert, P.; Kuroda, H.; Chino, N.; Watanabe, T.X.; Kimura, T.; Sakakibara, S.; et al. The cystine-stabilized alpha-helix: A common structural motif of ion-channel blocking neurotoxic peptides. Biopolymers 1991, 31, 1213–1220. [Google Scholar] [CrossRef]
- Ojeda, P.G.; Wang, C.K.; Craik, D.J. Chlorotoxin: Structure, activity, and potential uses in cancer therapy. Biopolymers 2016, 106, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Xiao, Y.; Kang, D.; Liu, J.; Li, Y.; Undheim, E.A.B.; Klint, J.K.; Rong, M.; Lai, R.; King, G.F. Discovery of a selective NaV1.7 inhibitor from centipede venom with analgesic efficacy exceeding morphine in rodent pain models. Proc. Natl. Acad. Sci. USA 2013, 110, 17534. [Google Scholar] [CrossRef] [PubMed]
- Lenhart, P.A.; Dash, S.T.; Mackay, W.P. A revision of the giant Amazonian ants of the genus Dinoponera (Hymenoptera, Formicidae). J. Hymenopt. Res. 2013, 31, 119–164. [Google Scholar] [CrossRef] [Green Version]
- Haddad Junior, V.; Cardoso, J.L.; Moraes, R.H. Description of an injury in a human caused by a false tocandira (Dinoponera gigantea, Perty, 1833) with a revision on folkloric, pharmacological and clinical aspects of the giant ants of the genera Paraponera and Dinoponera (sub-family Ponerinae). Rev. Inst. Med. Trop. Sao Paulo 2005, 47, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Madeira Jda, C.; Quinet, Y.P.; Nonato, D.T.; Sousa, P.L.; Chaves, E.M.; Jose Eduardo Ribeiro Honorio, J.; Pereira, M.G.; Assreuy, A.M. Novel Pharmacological Properties of Dinoponera quadriceps Giant Ant Venom. Nat. Prod. Commun. 2015, 10, 1607–1609. [Google Scholar] [CrossRef] [PubMed]
- Lopes, K.S.; Rios, E.R.; Lima, C.N.; Linhares, M.I.; Torres, A.F.; Havt, A.; Quinet, Y.P.; Fonteles, M.M.; Martins, A.M. The effects of the Brazilian ant Dinoponera quadriceps venom on chemically induced seizure models. Neurochem. Int. 2013, 63, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Noga, D.A.; Brandao, L.E.; Cagni, F.C.; Silva, D.; de Azevedo, D.L.; Araujo, A.; Dos Santos, W.F.; Miranda, A.; da Silva, R.H.; Ribeiro, A.M. Anticonvulsant Effects of Fractions Isolated from Dinoponera quadriceps (Kempt) Ant Venom (Formicidae: Ponerinae). Toxins (Basel) 2016, 9, 5. [Google Scholar] [CrossRef]
- Lima, D.B.; Torres, A.F.; Mello, C.P.; de Menezes, R.R.; Sampaio, T.L.; Canuto, J.A.; da Silva, J.J.; Freire, V.N.; Quinet, Y.P.; Havt, A.; et al. Antimicrobial effect of Dinoponera quadriceps (Hymenoptera: Formicidae) venom against Staphylococcus aureus strains. J. Appl. Microbiol. 2014, 117, 390–396. [Google Scholar] [CrossRef]
- Lima, D.B.; Sousa, P.L.; Torres, A.F.; Rodrigues, K.A.; Mello, C.P.; Menezes, R.R.; Tessarolo, L.D.; Quinet, Y.P.; de Oliveira, M.R.; Martins, A.M. Antiparasitic effect of Dinoponera quadriceps giant ant venom. Toxicon 2016, 120, 128–132. [Google Scholar] [CrossRef]
- Sousa, P.L.; Quinet, Y.; Ponte, E.L.; do Vale, J.F.; Torres, A.F.; Pereira, M.G.; Assreuy, A.M. Venom’s antinociceptive property in the primitive ant Dinoponera quadriceps. J. Ethnopharmacol. 2012, 144, 213–216. [Google Scholar] [CrossRef]
- Torres, A.F.; Huang, C.; Chong, C.M.; Leung, S.W.; Prieto-da-Silva, A.R.; Havt, A.; Quinet, Y.P.; Martins, A.M.; Lee, S.M.; Radis-Baptista, G. Transcriptome analysis in venom gland of the predatory giant ant Dinoponera quadriceps: Insights into the polypeptide toxin arsenal of hymenopterans. PLoS ONE 2014, 9, e87556. [Google Scholar] [CrossRef]
- Cologna, C.T.; Cardoso Jdos, S.; Jourdan, E.; Degueldre, M.; Upert, G.; Gilles, N.; Uetanabaro, A.P.; Costa Neto, E.M.; Thonart, P.; de Pauw, E.; et al. Peptidomic comparison and characterization of the major components of the venom of the giant ant Dinoponera quadriceps collected in four different areas of Brazil. J. Proteomics 2013, 94, 413–422. [Google Scholar] [CrossRef]
- Johnson, S.R.; Copello, J.A.; Evans, M.S.; Suarez, A.V. A biochemical characterization of the major peptides from the Venom of the giant Neotropical hunting ant Dinoponera australis. Toxicon 2010, 55, 702–710. [Google Scholar] [CrossRef]
- Hoffman, D.R. Ant venoms. Curr. Opin. Allergy Clin. Immunol. 2010, 10, 342–346. [Google Scholar] [CrossRef]
- Tholey, A.; Becker, A. Top-down proteomics for the analysis of proteolytic events—Methods, applications and perspectives. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2191–2199. [Google Scholar] [CrossRef]
- Blank, S.; Seismann, H.; Bockisch, B.; Braren, I.; Cifuentes, L.; McIntyre, M.; Ruhl, D.; Ring, J.; Bredehorst, R.; Ollert, M.W.; et al. Identification, recombinant expression, and characterization of the 100 kDa high molecular weight Hymenoptera venom allergens Api m 5 and Ves v 3. J. Immunol. 2010, 184, 5403–5413. [Google Scholar] [CrossRef]
- Tian, W.; Li, M.; Guo, H.; Peng, W.; Xue, X.; Hu, Y.; Liu, Y.; Zhao, Y.; Fang, X.; Wang, K.; et al. Architecture of the native major royal jelly protein 1 oligomer. Nat. Commun. 2018, 9, 3373. [Google Scholar] [CrossRef]
- Ciolek, J.; Reinfrank, H.; Quinton, L.; Viengchareun, S.; Stura, E.A.; Vera, L.; Sigismeau, S.; Mouillac, B.; Orcel, H.; Peigneur, S.; et al. Green mamba peptide targets type-2 vasopressin receptor against polycystic kidney disease. Proc. Natl. Acad. Sci. USA 2017, 114, 7154–7159. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Yu, H.; Yue, Y.; Liu, S.; Xing, R.; Chen, X.; Li, P. Combined proteomics and transcriptomics identifies sting-related toxins of jellyfish Cyanea nozakii. J. Proteomics 2016, 148, 57–64. [Google Scholar] [CrossRef]
- Leonardi, A.; Biass, D.; Kordis, D.; Stocklin, R.; Favreau, P.; Krizaj, I. Conus consors snail venom proteomics proposes functions, pathways, and novel families involved in its venomic system. J. Proteome Res. 2012, 11, 5046–5058. [Google Scholar] [CrossRef]
- Tasoulis, T.; Isbister, G.K. A Review and Database of Snake Venom Proteomes. Toxins (Basel) 2017, 9, 290. [Google Scholar] [CrossRef]
- Aili, S.R.; Touchard, A.; Petitclerc, F.; Dejean, A.; Orivel, J.; Padula, M.P.; Escoubas, P.; Nicholson, G.M. Combined Peptidomic and Proteomic Analysis of Electrically Stimulated and Manually Dissected Venom from the South American Bullet Ant Paraponera clavata. J. Proteome Res. 2017, 16, 1339–1351. [Google Scholar] [CrossRef]
- Santos, P.P.; Games, P.D.; Azevedo, D.O.; Barros, E.; de Oliveira, L.L.; de Oliveira Ramos, H.J.; Baracat-Pereira, M.C.; Serrao, J.E. Proteomic analysis of the venom of the predatory ant Pachycondyla striata (Hymenoptera: Formicidae). Arch. Insect Biochem. Physiol. 2017, 96. [Google Scholar] [CrossRef]
- Tani, N.; Kazuma, K.; Ohtsuka, Y.; Shigeri, Y.; Masuko, K.; Konno, K.; Inagaki, H. Mass Spectrometry Analysis and Biological Characterization of the Predatory Ant Odontomachus monticola Venom and Venom Sac Components. Toxins (Basel) 2019, 11, 50. [Google Scholar] [CrossRef]
- Aili, S.R.; Touchard, A.; Koh, J.M.; Dejean, A.; Orivel, J.; Padula, M.P.; Escoubas, P.; Nicholson, G.M. Comparisons of Protein and Peptide Complexity in Poneroid and Formicoid Ant Venoms. J. Proteome Res. 2016, 15, 3039–3054. [Google Scholar] [CrossRef]
- Zancolli, G.; Calvete, J.J.; Cardwell, M.D.; Greene, H.W.; Hayes, W.K.; Hegarty, M.J.; Herrmann, H.W.; Holycross, A.T.; Lannutti, D.I.; Mulley, J.F.; et al. When one phenotype is not enough: Divergent evolutionary trajectories govern venom variation in a widespread rattlesnake species. Proc. Biol. Sci. 2019, 286, 20182735. [Google Scholar] [CrossRef]
- Carcamo-Noriega, E.N.; Olamendi-Portugal, T.; Restano-Cassulini, R.; Rowe, A.; Uribe-Romero, S.J.; Becerril, B.; Possani, L.D. Intraspecific variation of Centruroides sculpturatus scorpion venom from two regions of Arizona. Arch. Biochem. Biophys. 2018, 638, 52–57. [Google Scholar] [CrossRef]
- Schaffrath, S.; Prendini, L.; Predel, R. Intraspecific venom variation in southern African scorpion species of the genera Parabuthus, Uroplectes and Opistophthalmus (Scorpiones: Buthidae, Scorpionidae). Toxicon 2018, 144, 83–90. [Google Scholar] [CrossRef]
- Schiener, M.; Hilger, C.; Eberlein, B.; Pascal, M.; Kuehn, A.; Revets, D.; Planchon, S.; Pietsch, G.; Serrano, P.; Moreno-Aguilar, C.; et al. The high molecular weight dipeptidyl peptidase IV Pol d 3 is a major allergen of Polistes dominula venom. Sci. Rep. 2018, 8, 1318. [Google Scholar] [CrossRef]
- Da Silva, J.R.; De Souza, A.Z.; Pirovani, C.P.; Costa, H.; Silva, A.; Dias, J.C.T.; Delabie, J.H.C.; Fontana, R. Assessing the Proteomic Activity of the Venom of the Ant Ectatomma tuberculatum (Hymenoptera: Formicidae: Ectatomminae). Psyche 2018, 2018, 11. [Google Scholar] [CrossRef]
- Kamakura, M.; Sakaki, T. A hypopharyngeal gland protein of the worker honeybee Apis mellifera L. enhances proliferation of primary-cultured rat hepatocytes and suppresses apoptosis in the absence of serum. Protein Expr. Purif. 2006, 45, 307–314. [Google Scholar] [CrossRef]
- Han, Q.; Fang, J.; Ding, H.; Johnson, J.K.; Christensen, B.M.; Li, J. Identification of Drosophila melanogaster yellow-f and yellow-f2 proteins as dopachrome-conversion enzymes. Biochem. J. 2002, 368, 333–340. [Google Scholar] [CrossRef]
- Fontana, R.; Mendes, M.; Souza, B.; Konno, K.; Marcondes César, L.M.; Malaspina, O.; Palma, M. Jelleines: A Family of Antimicrobial Peptides from the Royal Jelly of Honeybees (Apis mellifera). Peptides 2004, 25, 919–928. [Google Scholar] [CrossRef]
- Bolen, A.L.; Naren, A.P.; Yarlagadda, S.; Beranova-Giorgianni, S.; Chen, L.; Norman, D.; Baker, D.L.; Rowland, M.M.; Best, M.D.; Sano, T.; et al. The phospholipase A1 activity of lysophospholipase A-I links platelet activation to LPA production during blood coagulation. J. Lipid Res. 2011, 52, 958–970. [Google Scholar] [CrossRef] [Green Version]
- Hou, M.H.; Chuang, C.Y.; Ko, T.P.; Hu, N.J.; Chou, C.C.; Shih, Y.P.; Ho, C.L.; Wang, A.H. Crystal structure of vespid phospholipase A(1) reveals insights into the mechanism for cause of membrane dysfunction. Insect Biochem. Mol. Biol. 2016, 68, 79–88. [Google Scholar] [CrossRef]
- Rivera, R.; Chun, J. Biological effects of lysophospholipids. Rev. Physiol. Biochem. Pharmacol. 2008, 160, 25–46. [Google Scholar] [CrossRef]
- King, T.P.; Kochoumian, L.; Joslyn, A. Wasp venom proteins: Phospholipase A1 and B. Arch. Biochem. Biophys. 1984, 230, 1–12. [Google Scholar] [CrossRef]
- Yang, H.; Xu, X.; Ma, D.; Zhang, K.; Lai, R. A phospholipase A1 platelet activator from the wasp venom of Vespa magnifica (Smith). Toxicon 2008, 51, 289–296. [Google Scholar] [CrossRef]
- Perez-Riverol, A.; Lasa, A.M.; Dos Santos-Pinto, J.R.A.; Palma, M.S. Insect venom phospholipases A1 and A2: Roles in the envenoming process and allergy. Insect Biochem. Mol. Biol. 2019, 105, 10–24. [Google Scholar] [CrossRef]
- Lee, S.H.; Baek, J.H.; Yoon, K.A. Differential Properties of Venom Peptides and Proteins in Solitary vs. Social Hunting Wasps. Toxins 2016, 8, 32. [Google Scholar] [CrossRef]
- Kemeny, D.M.; Dalton, N.; Lawrence, A.J.; Pearce, F.L.; Vernon, C.A. The purification and characterisation of hyaluronidase from the venom of the honey bee, Apis mellifera. Eur. J. Biochem. 1984, 139, 217–223. [Google Scholar] [CrossRef]
- Gibbs, G.M.; Roelants, K.; O’Bryan, M.K. The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins—Roles in reproduction, cancer, and immune defense. Endocr. Rev. 2008, 29, 865–897. [Google Scholar] [CrossRef]
- Padavattan, S.; Schmidt, M.; Hoffman, D.; Marković-Housley, Z. Crystal Structure of the Major Allergen from Fire Ant Venom, Sol i 3. J. Mol. Biol. 2008, 383, 178–185. [Google Scholar] [CrossRef]
- Potiwat, R.; Sitcharungsi, R. Ant allergens and hypersensitivity reactions in response to ant stings. Asian Pac. J. Allergy Immunol. 2015, 33, 267–275. [Google Scholar]
- Borer, A.S.; Wassmann, P.; Schmidt, M.; Hoffman, D.R.; Zhou, J.J.; Wright, C.; Schirmer, T.; Markovic-Housley, Z. Crystal structure of Sol I 2: A major allergen from fire ant venom. J. Mol. Biol. 2012, 415, 635–648. [Google Scholar] [CrossRef]
- Lima, D.B.; Mello, C.P.; Bandeira, I.C.J.; Pessoa Bezerra de Menezes, R.R.P.; Sampaio, T.L.; Falcao, C.B.; Morlighem, J.R.L.; Radis-Baptista, G.; Martins, A.M.C. The dinoponeratoxin peptides from the giant ant Dinoponera quadriceps display in vitro antitrypanosomal activity. Biol. Chem. 2018, 399, 187–196. [Google Scholar] [CrossRef]
- Oldrati, V.; Koua, D.; Allard, P.M.; Hulo, N.; Arrell, M.; Nentwig, W.; Lisacek, F.; Wolfender, J.L.; Kuhn-Nentwig, L.; Stocklin, R. Peptidomic and transcriptomic profiling of four distinct spider venoms. PLoS ONE 2017, 12, e0172966. [Google Scholar] [CrossRef]
- Kalia, J.; Milescu, M.; Salvatierra, J.; Wagner, J.; Klint, J.K.; King, G.F.; Olivera, B.M.; Bosmans, F. From foe to friend: Using animal toxins to investigate ion channel function. J. Mol. Biol. 2015, 427, 158–175. [Google Scholar] [CrossRef]
- Koehbach, J. Structure-Activity Relationships of Insect Defensins. Front. Chem. 2017, 5, 45. [Google Scholar] [CrossRef]
- Zhang, M.; Fishman, Y.; Sher, D.; Zlotkin, E. Hydralysin, a novel animal group-selective paralytic and cytolytic protein from a noncnidocystic origin in hydra. Biochemistry 2003, 42, 8939–8944. [Google Scholar] [CrossRef]
- Sher, D.; Fishman, Y.; Zhang, M.; Lebendiker, M.; Gaathon, A.; Mancheno, J.M.; Zlotkin, E. Hydralysins, a new category of beta-pore-forming toxins in cnidaria. J. Biol. Chem. 2005, 280, 22847–22855. [Google Scholar] [CrossRef]
- Monnin, T.; Peeters, C. Monogyny and regulation of worker mating in the queenless ant Dinoponera quadriceps. Anim. Behav. 1998, 55, 299–306. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Westermeier, R.; Naven, T.; Höpker, H.-R. Proteomics in Practice: A Guide to Successful Experimental Design, 2nd ed.; Wiley-VCH Verlag GmbH & Co.: Weinheim, Germany, 2008. [Google Scholar] [CrossRef]
- Beraldo Neto, E.; Mariano, D.O.C.; Freitas, L.A.; Dorce, A.L.C.; Martins, A.N.; Pimenta, D.C.; Portaro, F.C.V.; Cajado-Carvalho, D.; Dorce, V.A.C.; Nencioni, A.L.A. Tb II-I, a Fraction Isolated from Tityus bahiensis Scorpion Venom, Alters Cytokines’: Level and Induces Seizures When Intrahippocampally Injected in Rats. Toxins (Basel) 2018, 10, 250. [Google Scholar] [CrossRef]
- Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. PEAKS: Powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 2337–2342. [Google Scholar] [CrossRef]
- Patalano, S.; Vlasova, A.; Wyatt, C.; Ewels, P.; Camara, F.; Ferreira, P.G.; Asher, C.L.; Jurkowski, T.P.; Segonds-Pichon, A.; Bachman, M.; et al. Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. Proc. Natl. Acad. Sci. USA 2015, 112, 13970–13975. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]











© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ceolin Mariano, D.O.; de Oliveira, Ú.C.; Zaharenko, A.J.; Pimenta, D.C.; Rádis-Baptista, G.; Prieto-da-Silva, Á.R.d.B. Bottom-Up Proteomic Analysis of Polypeptide Venom Components of the Giant Ant Dinoponera Quadriceps. Toxins 2019, 11, 448. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11080448
Ceolin Mariano DO, de Oliveira ÚC, Zaharenko AJ, Pimenta DC, Rádis-Baptista G, Prieto-da-Silva ÁRdB. Bottom-Up Proteomic Analysis of Polypeptide Venom Components of the Giant Ant Dinoponera Quadriceps. Toxins. 2019; 11(8):448. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11080448
Chicago/Turabian StyleCeolin Mariano, Douglas Oscar, Úrsula Castro de Oliveira, André Junqueira Zaharenko, Daniel Carvalho Pimenta, Gandhi Rádis-Baptista, and Álvaro Rossan de Brandão Prieto-da-Silva. 2019. "Bottom-Up Proteomic Analysis of Polypeptide Venom Components of the Giant Ant Dinoponera Quadriceps" Toxins 11, no. 8: 448. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11080448