The Cadherin Cry1Ac Binding-Region is Necessary for the Cooperative Effect with ABCC2 Transporter Enhancing Insecticidal Activity of Bacillus thuringiensis Cry1Ac Toxin
Abstract
:1. Introduction
2. Results
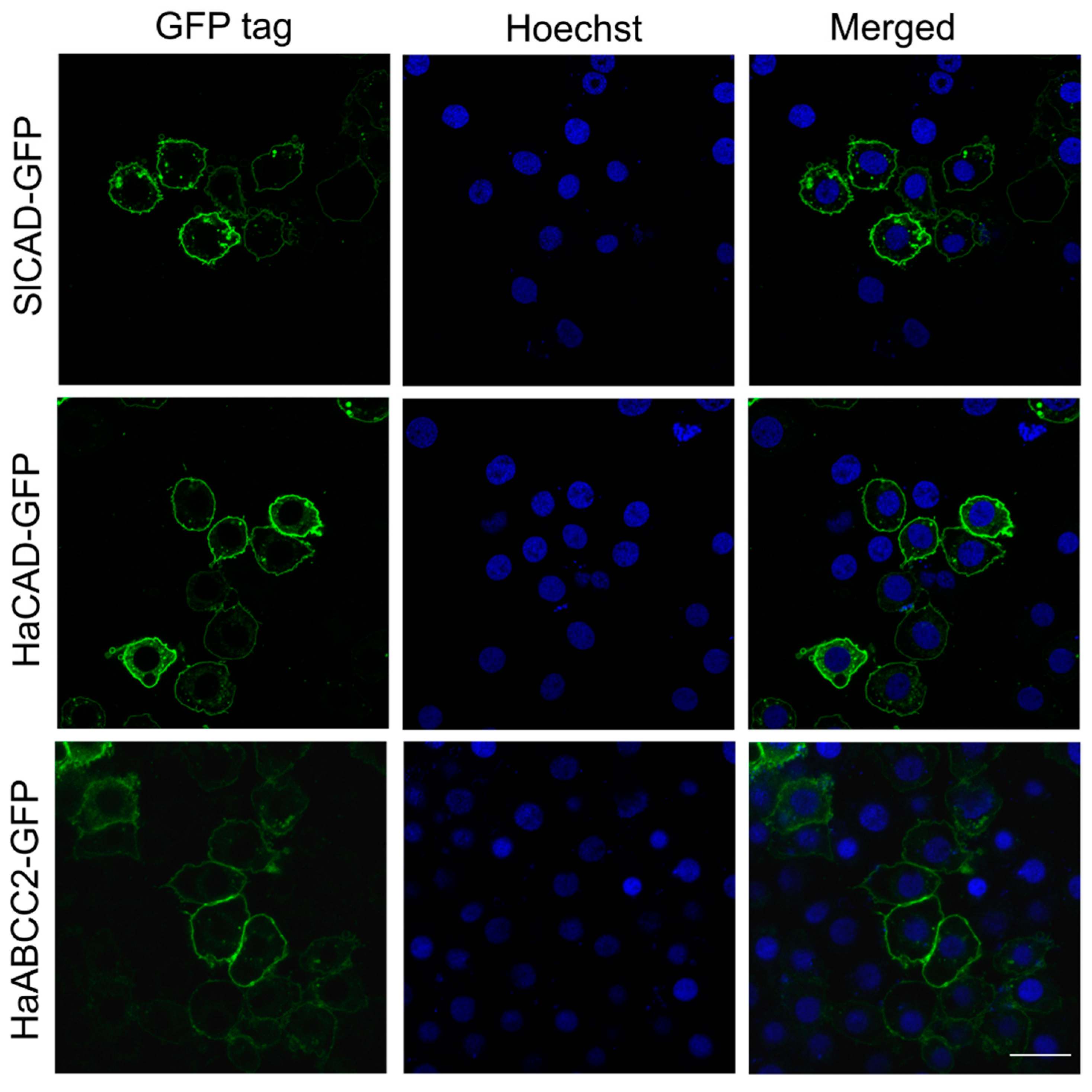
2.1. HaCAD-GFP Mediates Cytotoxicity of Cry1Ac in Hi5 Cells in Contrast to SlCAD-GFP
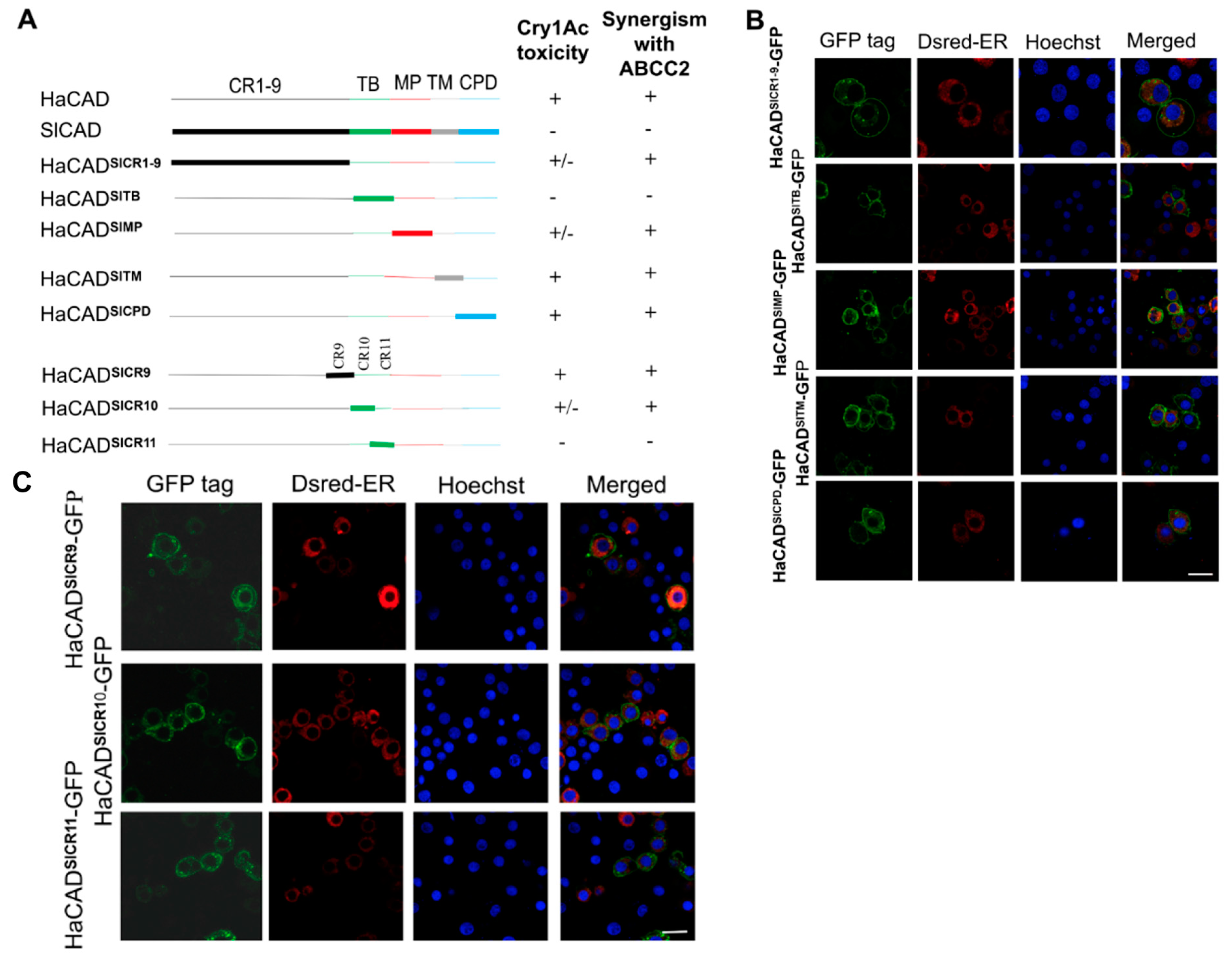
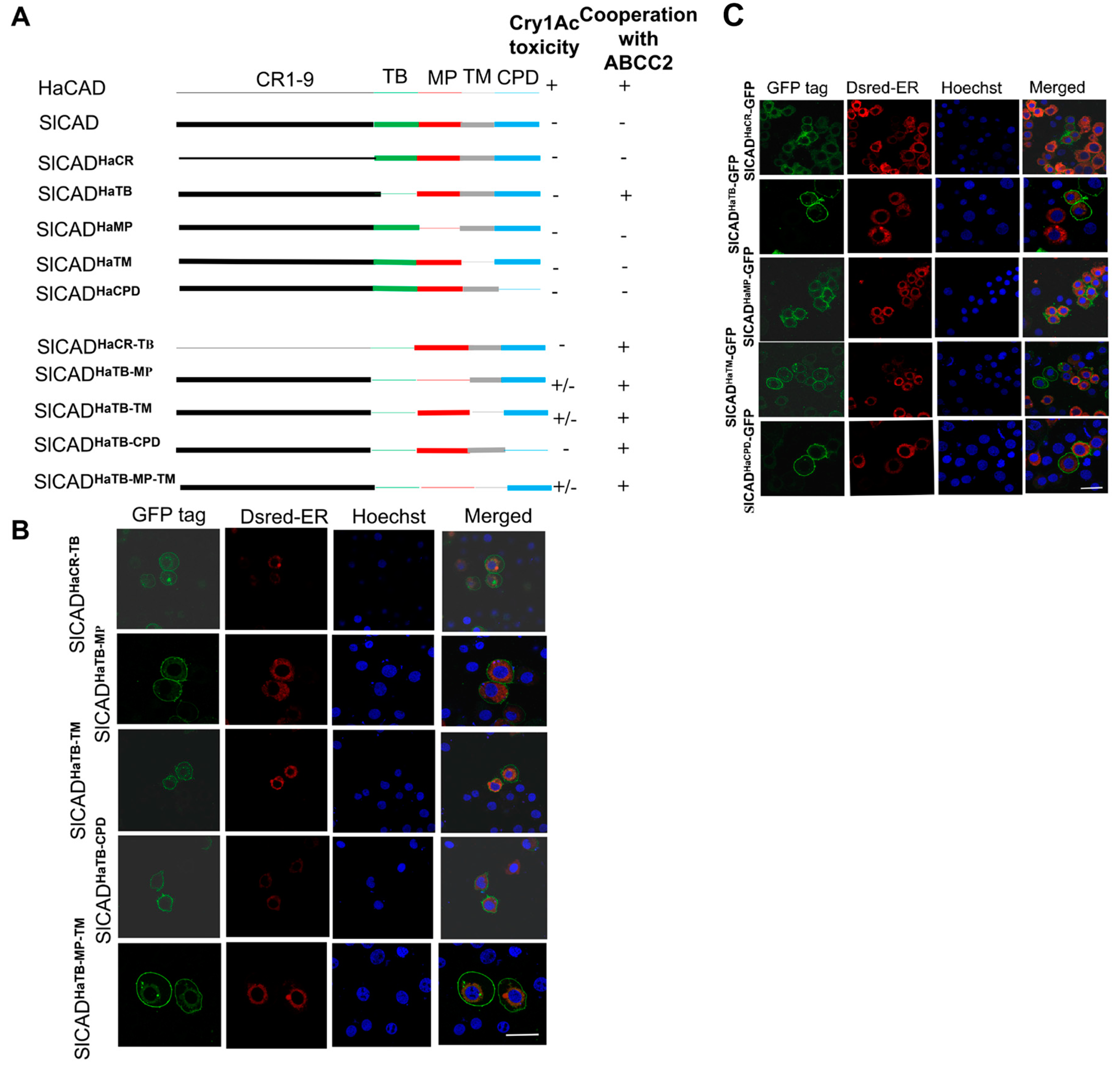
2.2. The HaCAD TB Domain is Necessary to Cooperate with HaABCC2 Resulting in High Cry1Ac Cytotoxicity
2.3. Additional Regions Besides TB Region are Necessary to Induce Toxicity of Cry1Ac When ABCC2 is Absent
2.4. The TB Fragment from HaCAD-GFP Binds Cry1Ac and TB Fragment Inhibits Cry1Ac Cytotoxicity in Hi5 Cells Transfected with HaCAD-GFP
2.5. The CPD Region of HaCAD-GFP is not Necessary to Mediate Cry1Ac Toxicity neither to Synergize the Toxicity of Cry1Ac with ABCC2
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cry1Ac Toxin
4.2. Cloning of SlCAD
4.3. Plasmids for Protein Expression in Hi5 Insect Cells
4.4. Expression and Purification of Proteins in E. coli
4.5. Transfection
4.6. Microscopic Observation
4.7. Cytotoxicity Assay
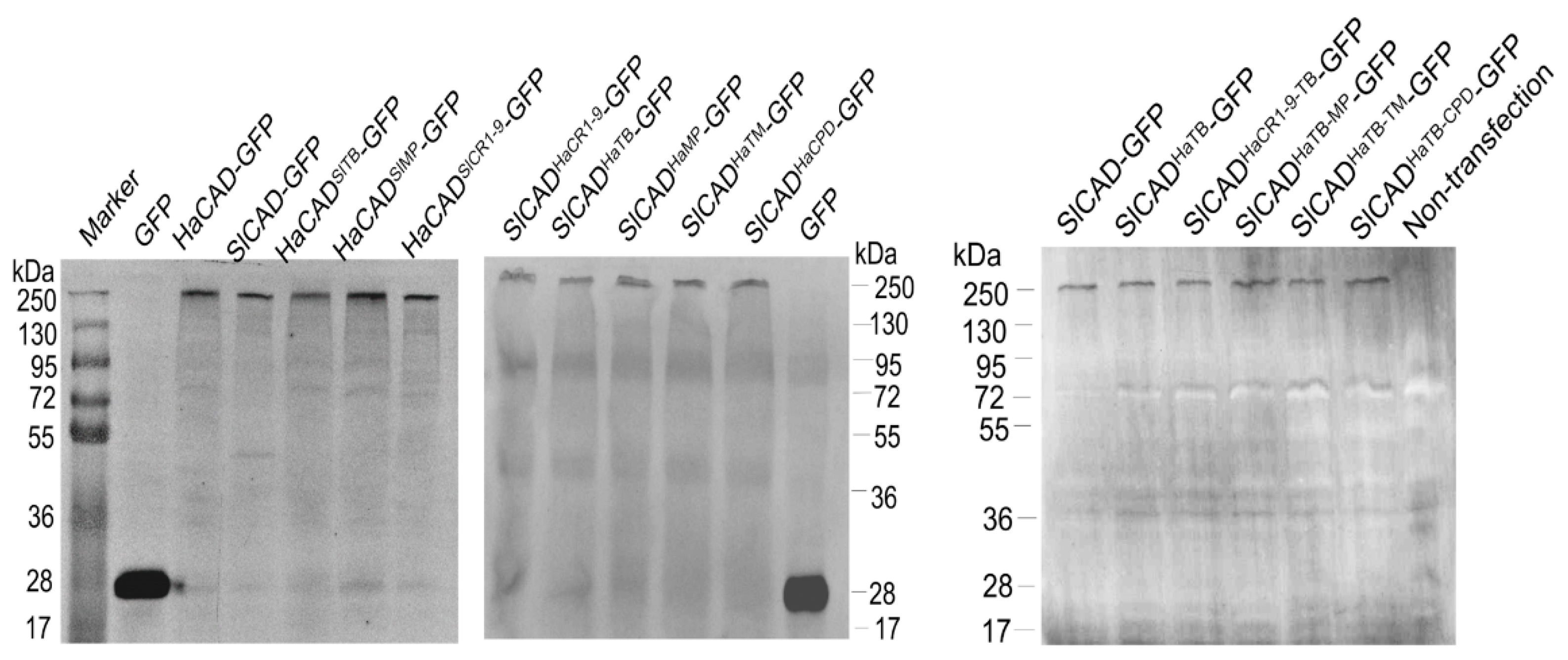
4.8. Western Blot Assay
4.9. Pull-down Assay
4.10. Real Time RT-qPCR Assay
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lu, Y.; Wu, K.; Jiang, Y.; Guo, Y.; Desneux, N. Widespread adoption of Bt cotton and insecticide decrease promotes biocontrol services. Nature 2012, 487, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Zhang, H.; Lu, Y.; Yang, Y.; Wu, K.; Tabashnik, B.E.; Wu, Y. Large-scale test of the natural refuge strategy for delaying insect resistance to transgenic Bt crops. Nat. Biotechnol. 2015, 33, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Gahan, L.J.; Gould, F.; Heckel, D.G. Identification of a gene associated with Bt resistance in Heliothis virescens. Science 2001, 293, 857–860. [Google Scholar] [CrossRef] [PubMed]
- Gahan, L.J.; Pauchet, Y.; Vogel, H.; Heckel, D.G. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 2010, 6, e1001248. [Google Scholar] [CrossRef] [PubMed]
- Heckel, D.G. Learning the ABCs of Bt: ABC transporters and insect resistance to Bacillus thuringiensis provide clues to a crucial step in toxin mode of action. Pestic. Biochem. Physiol. 2012, 104, 103–110. [Google Scholar] [CrossRef]
- Gómez, I.; Dean, D.H.; Bravo, A.; Soberón, M. Molecular basis for Bacillus thuringiensis Cry1Ab toxin specificity: Two structural determinants in the Manduca sexta Bt-R1 receptor interact with loops α-8 and 2 in domain II of Cy1Ab toxin. Biochemistry 2003, 42, c10482–c10489. [Google Scholar] [CrossRef] [PubMed]
- Ocelot, J.; Sánchez, J.; Gómez, I.; Tabashnik, B.E.; Bravo, A.; Soberón, M. ABCC2 is associated with Bacillus thuringiensis Cry1Ac toxin oligomerization and membrane insertion in diamondback moth. Sci. Rep. 2017, 7, 2386. [Google Scholar] [CrossRef] [PubMed]
- Bravo, A.; Likitvivatanavong, S.; Gill, S.S. Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 2011, 41, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Pardo-López, L.; Gómez, I.; Rausell, C.; Sánchez, J.; Soberón, M.; Bravo, A. Structural changes of the Cry1Ac oligomeric pre-pore from Bacillus thuringiensis induced by N-acetylgalactosamine facilitates toxin membrane insertion. Biochemistry 2006, 45, 10329–10336. [Google Scholar] [CrossRef]
- Arenas, I.; Bravo, A.; Soberón, M.; Gómez, I. Role of alkaline phosphatase from Manduca sexta in the mechanism of action of Bacillus thuringiensis Cry1Ab toxin. J. Biol. Chem. 2010, 285, 12497–12503. [Google Scholar] [CrossRef]
- Tanaka, S.; Miyamoto, K.; Noda, H.; Jurat-Fuentes, J.L.; Yoshizawa, Y.; Endo, H.; Sato, R. The ATP-binding cassette transporter subfamily C member 2 in Bombyx mori larvae is a functional receptor for Cry toxins from Bacillus thuringiensis. FEBS. J 2013, 280, 1782–1794. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Miyamoto, K.; Noda, H.; Endo, H.; Kikuta, S.; Sato, R. Single amino acid insertions in extracellular loop 2 of Bombyx mori ABCC2 disrupt its receptor function for Bacillus thuringiensis Cry1Ab and Cry1Ac but not Cry1Aa toxins. Peptides 2016, 78, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; He, F.; Xiao, Y.; Liu, C.; Li, J.; Yang, Y.; Ai, H.; Peng, J.; Hong, H.; Liu, K. Endogenous expression of a Bt toxin receptor in the Cry1Ac-susceptible insect cell line and its synergistic effect with cadherin on cytotoxicity of activated Cry1Ac. Insect Biochem. Mol. Biol. 2015, 59, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Bretschneider, A.; Heckel, D.G.; Pauchet, Y. Three toxins, two receptors, one mechanism: Mode of action of Cry1A toxins from Bacillus thuringiensis in Heliothis virescens. Insect Biochem. Mol. Biol. 2016, 76, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.L.; Jiang, W.L.; Ma, Y.J.; Hu, H.Y.; Ma, X.Y.; Ma, Y.; Li, G.Q. The Spodoptera exigua (Lepidoptera: Noctuidae) ABCC2 mediates Cry1Ac cytotoxicity and in conjunction with cadherin contributes to enhance Cry1Ca toxicity in Sf9 cells. J. Econ. Entomol. 2016, 109, 2281–2289. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wu, K.; Liang, G.; Guo, Y. Gene cloning and expression of cadherin in midgut of Helicoverpa armigera and its Cry1A binding region. Sci. China C Life. Sci 2005, 48, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, H.; Wu, S.; Yang, Y.; Xu, X.; Wu, Y. Identification and molecular detection of a deletion mutation responsible for a truncated cadherin of Helicoverpa armigera. Insect Biochem. Mol. Biol. 2006, 36, 735–740. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, S.; Shi, Y.; Yang, Y.; Fabrick, J.A.; Wu, Y. Intra-and extracellular domains of the Helicoverpa armigera cadherin mediate Cry1Ac cytotoxicity. Insect Biochem. Mol. Biol. 2017, 86, 41–49. [Google Scholar] [CrossRef]
- Zhao, J.; Jin, L.; Yang, Y.; Wu, Y. Diverse cadherin mutations conferring resistance to Bacillus thuringiensis toxin Cry1Ac in Helicoverpa armigera. Insect Biochem. Mol. Biol. 2010, 40, 113–118. [Google Scholar] [CrossRef]
- Fabrick, J.A.; Tabashnik, B.E. Binding of Bacillus thuringiensis toxin Cry1Ac to multiple sites of cadherin in pink bollworm. Insect Biochem. Mol. Biol. 2007, 37, 97–106. [Google Scholar] [CrossRef]
- Dorsch, J.A.; Candas, M.; Griko, N.B.; Maaty, W.S.; Midboe, E.G.; Vadlamudi, R.K.; Bulla, L.A., Jr. Cry1A toxins of Bacillus thuringiensis bind specifically to a region adjacent to the membrane-proximal extracellular domain of BT-R1 in Manduca sexta: Involvement of a cadherin in the entomopathogenicity of Bacillus thuringiensis. Insect Biochem. Mol. Biol. 2002, 32, 1025–1036. [Google Scholar] [CrossRef]
- Pacheco, S.; Gómez, I.; Gill, S.S.; Bravo, A.; Soberón, M. Enhancement of insecticidal activity of Bacillus thuringiensis Cry1A toxins by fragments of a toxin-binding cadherin correlates with oligomer formation. Peptides 2009, 30, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, M.A.; Moussa, S.; Taylor, M.D. Manduca sexta (Lepidoptera: Sphingidae) cadherin fragments function as synergists for Cry1A and Cry1C Bacillus thuringiensis toxins against noctuid moths Helicoverpa zea, Agrotis ipsilon and Spodoptera exigua. Pest Manag. Sci. 2009, 65, 1097–1103. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Adegawa, S.; Kikuta, S.; Sato, R. The intracellular region of silkworm cadherin like protein is not necessary to mediate the toxicity of Bacillus thuringiensis Cry1Aa and Cry1Ab toxins. Insect Biochem. Mol. Biol. 2018, 94, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Jurat-Fuentes, J.L.; Adang, M.J. The Heliothis virescens cadherin protein expressed in Drosophila S2 cells functions as a receptor for Bacillus thuringiensis Cry1A but not Cry1Fa toxins. Biochemistry 2006, 45, 9688–9695. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Endo, H.; Adegawa, S.; Kikuta, S.; Sato, R. Functional characterization of Bacillus thuringiensis Cry toxin receptors explain resistance in insects. FEBS. J. 2016, 283, 4474–4490. [Google Scholar] [CrossRef] [PubMed]
- Gómez, I.; Sánchez, J.; Miranda, R.; Bravo, A. Cadherin-like receptor binding facilitates proteolytic cleavage of helix α-1 in domain I and oligomer pre-pore formation of Bacillus thuringiensis Cry1Ab toxin. FEBS Lett. 2002, 513, 242–246. [Google Scholar] [CrossRef]
- Hua, G.; Jurat-Fuentes, J.L.; Adang, M.J. Bt-R1 a extracellular cadherin repeat 12 mediates Bacillus thuringiensis Cry1Ab binding and cytotoxicity. J. Biol. Chem. 2004, 279, 28051–28056. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Solís, M.; Pinos, D.; Endo, H.; Portugal, L.; Sato, R.; Ferré, J.; Herrero, S.; Hernández-Martínez, P. Role of Bacillus thuringiensis Cry1A toxins domains in the binding to the ABCC2 receptor from Spodoptera exigua. Insect Biochem. Mol. Biol. 2018, 101, 47–56. [Google Scholar] [CrossRef]
- Chen, J.; Hua, G.; Jurat-Fuentes, J.L. Synergism of Bacillus thuringiensis toxins by a fragment of a toxin-binding cadherin. Proc. Natl. Acad. Sci. USA 2007, 104, 13901–13916. [Google Scholar] [CrossRef]
- Rahman, K.; Abdullah, M.A.F.; Ambati, S.; Taylor, M.D.; Adang, M.J. Differential protection of cry1Fa toxin against Spodoptera frugiperda larval gut proteases by cadherin orthologs correlates with increased synergism. Appl. Environ. Microbiol. 2012, 78, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Xu, X.; Ye, W.; Yu, Z.; Sun, M. Helicoverpa armigera cadherin fragment enhances Cry1Ac insecticidal activity by facilitating toxin-oligomer formation. Appl. Microbiol. Biotechnol. 2010, 85, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Candas, M.; Grinko, N.B.; Taussig, R.; Bulla, L.A. A mechanism of cell death involving an adenylyl cyclase/PKA-signaling pathway is induced by the Cry1Ab toxin of Bacillus thuringiensis. Proc. Natl. Acad. Sci. USA 2006, 103, 9897–9902. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, S.; Yang, Y.; Tabashnik, B.E.; Wu, Y. Non-recessive Bt toxin resistance conferred by an intracellular cadherin mutation in field-selected populations of cotton bollworm. PLoS ONE 2012, 7, e53418. [Google Scholar] [CrossRef] [PubMed]
- Pusztai-Carey, M.; Carey, P.; Lessard, T.; Yaguchi, M. Isolation, Quantitation and Purification of Insecticidal Proteins from Bacillus thuringiensis. US5356788A, 18 October 1994. [Google Scholar]
- Xu, P.; Islam, M.; Xiao, Y.; He, F.; Li, Y.; Peng, J.; Hong, H.; Liu, C.; Liu, K. Expression of recombinant and mosaic Cry1Ac receptors from Helicoverpa armigera and their influences on the cytotoxicity of activated Cry1Ac to Spodoptera litura Sl-HP cells. Cytotechnology 2016, 68, 481–496. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, K.; Zhang, D.; Gong, L.; He, F.; Soberón, M.; Bravo, A.; Tabashnik, B.E.; Wu, K. Resistance to Bacillus thuringiensis mediated by an ABC transporter mutation increases susceptibility to toxins from other bacteria in an invasive insect. PLoS Pathog. 2016, 12, e1005450. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Dai, Q.; Hu, R.; Pacheco, S.; Yang, Y.; Liang, G.; Soberón, M.; Bravo, A.; Liu, K.; Wu, K. A single point mutation resulting in cadherin mis-localization underpins resistance against Bacillus thuringiensis toxin in cotton bollworm. J. Biol. Chem. 2017, 292, 2933–2943. [Google Scholar] [CrossRef]
- Liu, L.; Chen, Z.; Yang, Y.; Xiao, Y.; Liu, C.; Ma, Y.; Soberón, M.; Bravo, A.; Yang, Y.; Liu, K. A single amino acid polymorphism in ABCC2 loop 1 is responsible for differential toxicity of Bacillus thuringiensis Cry1Ac toxin in different Spodoptera (Noctuidae) species. Insect Biochem. Mol. Biol. 2018, 100, 59–65. [Google Scholar] [CrossRef]
- Su, P.; Guan, H.; Zhang, Y.; Wang, X.; Gao, L.; Zhao, Y.; Hu, T.; Zhou, J.; Ma, B.; Tu, L.; et al. Probing the single key amino acid responsible for the novel catalytic function of ent-kaurene oxidase supported by NADPH-cytochrome P450 reductases in Tripterygium wilfordii. Front. Plant Sci. 2017, 8, 1756. [Google Scholar] [CrossRef]
- Lin, Y.; Nomura, T.; Cheong, J.H.; Dorjsuren, D.; Iida, K.; Murakami, S. Hepatitis B virus X protein is a transcriptional modulator that communicates with transcriptional factor IIB and RNA polymerase II subunit 5. J. Biol. Chem. 1997, 272, 7132–7139. [Google Scholar] [CrossRef]
- Wei, J.; Liang, G.; Wu, K.; Gu, S.; Guo, Y.; Ni, X.; Li, X. Cytotoxicity and binding profiles of activated Cry1Ac and Cry2Ab to three insect cell lines. Insect Sci. 2018, 25, 655–666. [Google Scholar] [CrossRef] [PubMed]
| Protein | EC50 (μg/mL) | 95% CI (μg/mL) | Slope | x2 | df | Susceptibility |
|---|---|---|---|---|---|---|
| SlCAD-GFP | >40 a | N | N | N | N | − |
| HaCAD-GFP | 7.36 | 6.23–8.59 | 3.11 | 2.90 | 3 | + |
| HaABCC2-GFP | 0.26 | 0.15–0.44 | 4.09 | 13.27 | 3 | + |
| Protein | EC50 (µg/mL) | 95% CI (µg/mL) | Slope | x2 | df | Potentiation of Cry1Ac toxicity |
|---|---|---|---|---|---|---|
| GFP (control) a | >40 a | N | N | N | N | N |
| HaABCC2-GFP | 0.26 | 0.15–0.44 | 4.09 | 13.27 | 3 | N |
| GFP+HaABCC2-GFP | 0.28 | 0.23–0.34 | 3.00 | 0.63 | 3 | − |
| SlCAD-GFP+HaABCC2-GFP | 0.28 | 0.24–0.35 | 3.03 | 1.16 | 3 | − |
| HaCAD-GFP+HaABCC2-GFP | 0.01 | 0.01–0.02 | 3.33 | 11.17 | 3 | + (28 fold) |
| HaCAD-GFPSlCR1-9+HaABCC2-GFP | 0.02 | 0.02–0.03 | 3.24 | 1.54 | 3 | + (14 fold) |
| HaCAD-GFPSlTB+HaABCC2-GFP | 0.28 | 0.24–0.33 | 3.47 | 1.81 | 3 | − |
| HaCAD-GFPSlMP+HaABCC2-GFP | 0.01 | 0.01–0.03 | 2.99 | 8.81 | 3 | + (28 fold) |
| HaCAD-GFPSlTM+HaABCC2-GFP | 0.01 | 0.004–0.02 | 3.30 | 8.6 | 3 | + (28 fold) |
| HaCAD-GFPSlCPD+HaABCC2-GFP | 0.02 | 0.01–0.02 | 3.11 | 2.39 | 3 | + (14 fold) |
| HaCAD-GFPSlCR9+HaABCC2-GFP | 0.02 | 0.01–0.04 | 2.82 | 8.99 | 3 | + (14 fold) |
| HaCAD-GFPSlCR10+HaABCC2-GFP | 0.03 | 0.02–0.03 | 2.70 | 3.44 | 3 | + (9.3 fold) |
| HaCAD-GFPSlCR11+HaABCC2-GFP | 0.20 | 0.09–0.39 | 3.35 | 14.87 | 3 | − |
| SlCAD-GFPHaCR1-9+HaABCC2-GFP | 0.20 | 0.14–0.29 | 4.22 | 7.61 | 3 | − |
| SlCAD-GFPHaTB+HaABCC2-GFP | 0.01 | 0.01–0.01 | 5.11 | 3.31 | 3 | + (28 fold) |
| SlCAD-GFPHaMP+HaABCC2-GFP | 0.21 | 0.14–0.29 | 4.41 | 7.35 | 3 | − |
| SlCAD-GFPHaTM+HaABCC2-GFP | 0.30 | 0.19–0.47 | 5.18 | 12.74 | 3 | − |
| SlCAD-GFPHaCPD+HaABCC2-GFP | 0.20 | 0.18–0.22 | 5.02 | 3.75 | 3 | − |
| SlCAD-GFPHaCR1-9,TB+HaABCC2-GFP | 0.01 | 0.005–0.02 | 3.00 | 6.26 | 3 | +(28 fold) |
| SlCAD-GFP HaTB, MP+HaABCC2-GFP | 0.01 | 0.01–0.02 | 4.42 | 4.31 | 3 | + (28 fold) |
| SlCAD-GFPHevTB + GFP a | >40 a | N | N | N | N | N |
| SlCAD-GFPHevTB+HaABCC2-GFP | 0.02 | 0.02–0.03 | 5.77 | 3.62 | 3 | + (14 fold) |
| Protein | % cell swelling at 40 μg/mL | EC50 (μg/mL) | 95% CI (μg/mL) | Slope | x2 | df | Susceptibility |
|---|---|---|---|---|---|---|---|
| GFP (control) a | 2.11 ± 1.17 | N | N | N | N | N | − |
| SlCAD-GFP a | 2.13 ± 0.32 | N | N | N | N | N | − |
| HaCAD-GFP | 96.23 ± 3.66 | 7.36 | 6.23–8.59 | 3.11 | 2.90 | 3 | + |
| HaCAD-GFPSlCR1-9 | 44.66 ± 1.15 | N | N | N | N | N | +/− |
| HaCAD-GFPSlTBa | 1.30 ± 0.53 | N | N | N | N | N | − |
| HaCAD-GFPSlMP | 50.00 ± 4.58 | N | N | N | N | N | +/− |
| HaCAD-GFPSlTM | 92.83 ± 1.41 | 13.12 | 11.62–14.87 | 4.48 | 0.249 | 3 | + |
| HaCAD-GFPSlCPD | 81.64 + 6.41 | 8.65 | 7.35–10.13 | 3.03 | 3.65 | 3 | + |
| HaCAD-GFPSlCR9 | 81.92 ± 9.34 | 10.78 | 9.32–12.5 | 3.40 | 3.03 | 3 | + |
| HaCAD-GFPSlCR10 | 16.07 ± 4.70 | N | N | N | N | N | +/− |
| HaCAD-GFPSlCR11 | 6.23 ± 1.68 | N | N | N | N | N | − |
| SlCAD-GFPHaCRa | 1.95 ± 0.82 | N | N | N | N | N | − |
| SlCAD-GFPHaTBa | 4.73 ± 0.70 | N | N | N | N | N | − |
| SlCAD-GFPHaMPa | 2.86 ± 0.47 | N | N | N | N | N | − |
| SlCAD-GFPHaTMa | 4.91 ± 0.75 | N | N | N | N | N | − |
| SlCAD-GFPHaCPDa | 1.64 ± 0.55 | N | N | N | N | N | − |
| SlCAD-GFPHevTBa | No cell swellinga | N | N | N | N | N | − |
| Protein | % Cell Swelling at 40 μg/mL | Susceptibility |
|---|---|---|
| SlCAD-GFPHaTB a | 1.33 ± 0.57 | − |
| SlCAD-GFPHaTB, MP | 48.60 ± 1.79 | +/− |
| SlCAD-GFPHaTB, TM | 46.31 ± 3.96 | +/− |
| SlCAD-GFPHaTB, CPD a | 4.71 ± 3.01 | − |
| SlCAD-GFPHaCR-TB, TM | 42.19 ± 3.97 | +/− |
| SlCAD-GFPHaTB, MP, TM | 52.87 ± 1.86 | +/− |
| Mixture | EC50 (µg/mL) | 95% of CI (µg/mL) | Slope | x2 | df | Inhibition |
|---|---|---|---|---|---|---|
| Cry1Ac+GST | 12.96 | 7.75–23.14 | 4.38 | 14.51 | 3 | − |
| Cry1Ac+GST-TB-MPHaCAD | >40 a | N | N | N | N | + |
| Cry1Ac+GST-TB-MPSlCAD | 15.38 | 13.21–18.12 | 3.25 | 2.53 | 3 | − |
| Cry1Ac+GST-MPHaCAD | 14.35 | 12.60–16.43 | 4.07 | 0.61 | 3 | − |
| Cry1Ac+GST-TBHaCAD | >40 a | N | N | N | N | + |
| Mixture | Expressed Receptor in Hi5 cells | EC50 (µg/mL) | 95% of CI (µg/mL) | Slope | x2 | df | Inhibition |
|---|---|---|---|---|---|---|---|
| Cry1Ac+GST | HaABCC2-GFP | 0.15 | 0.07–0.25 | 3.15 | 11.04 | 3 | − |
| Cry1Ac+GST | HaCAD-GFP+HaABCC2-GFP | 0.013 | 0.010–0.016 | 2.39 | 3.32 | 3 | − |
| Cry1Ac+GST-TBHaCAD | HaABCC2-GFP | 0.11 | 0.01–0.21 | 2.62 | 15.12 | 3 | − |
| Cry1Ac+GST-TBHaCAD | HaCAD-GFP+HaABCC2-GFP | 0.02 | 0.01–0.03 | 2.30 | 14.61 | 3 | − |
| Cry1Ac+GST-TB-MPHaCAD | HaABCC2-GFP | 0.15 | 0.06–0.28 | 3.12 | 13.96 | 3 | − |
| Cry1Ac+GST-TB-MPHaCAD | HaCAD-GFP+HaABCC2-GFP | 0.014 | 0.005–0.028 | 2.37 | 10.51 | 3 | − |
| Cry1Ac+GST-MPHaCAD | HaABCC2-GFP | 0.19 | 0.06–0.44 | 3.54 | 21.68 | 3 | − |
| Cry1Ac+GST-MPHaCAD | HaCAD-GFP+HaABCC2-GFP | 0.013 | 0.011–0.015 | 3.96 | 3.66 | 3 | − |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Zhang, J.; Xiao, Y.; Yang, Y.; Liu, C.; Peng, R.; Yang, Y.; Bravo, A.; Soberón, M.; Liu, K. The Cadherin Cry1Ac Binding-Region is Necessary for the Cooperative Effect with ABCC2 Transporter Enhancing Insecticidal Activity of Bacillus thuringiensis Cry1Ac Toxin. Toxins 2019, 11, 538. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11090538
Ma Y, Zhang J, Xiao Y, Yang Y, Liu C, Peng R, Yang Y, Bravo A, Soberón M, Liu K. The Cadherin Cry1Ac Binding-Region is Necessary for the Cooperative Effect with ABCC2 Transporter Enhancing Insecticidal Activity of Bacillus thuringiensis Cry1Ac Toxin. Toxins. 2019; 11(9):538. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11090538
Chicago/Turabian StyleMa, Yuemin, Jianfeng Zhang, Yutao Xiao, Yanchao Yang, Chenxi Liu, Rong Peng, Yongbo Yang, Alejandra Bravo, Mario Soberón, and Kaiyu Liu. 2019. "The Cadherin Cry1Ac Binding-Region is Necessary for the Cooperative Effect with ABCC2 Transporter Enhancing Insecticidal Activity of Bacillus thuringiensis Cry1Ac Toxin" Toxins 11, no. 9: 538. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11090538






