Widespread Evolution of Molecular Resistance to Snake Venom α-Neurotoxins in Vertebrates
Abstract
:1. Introduction
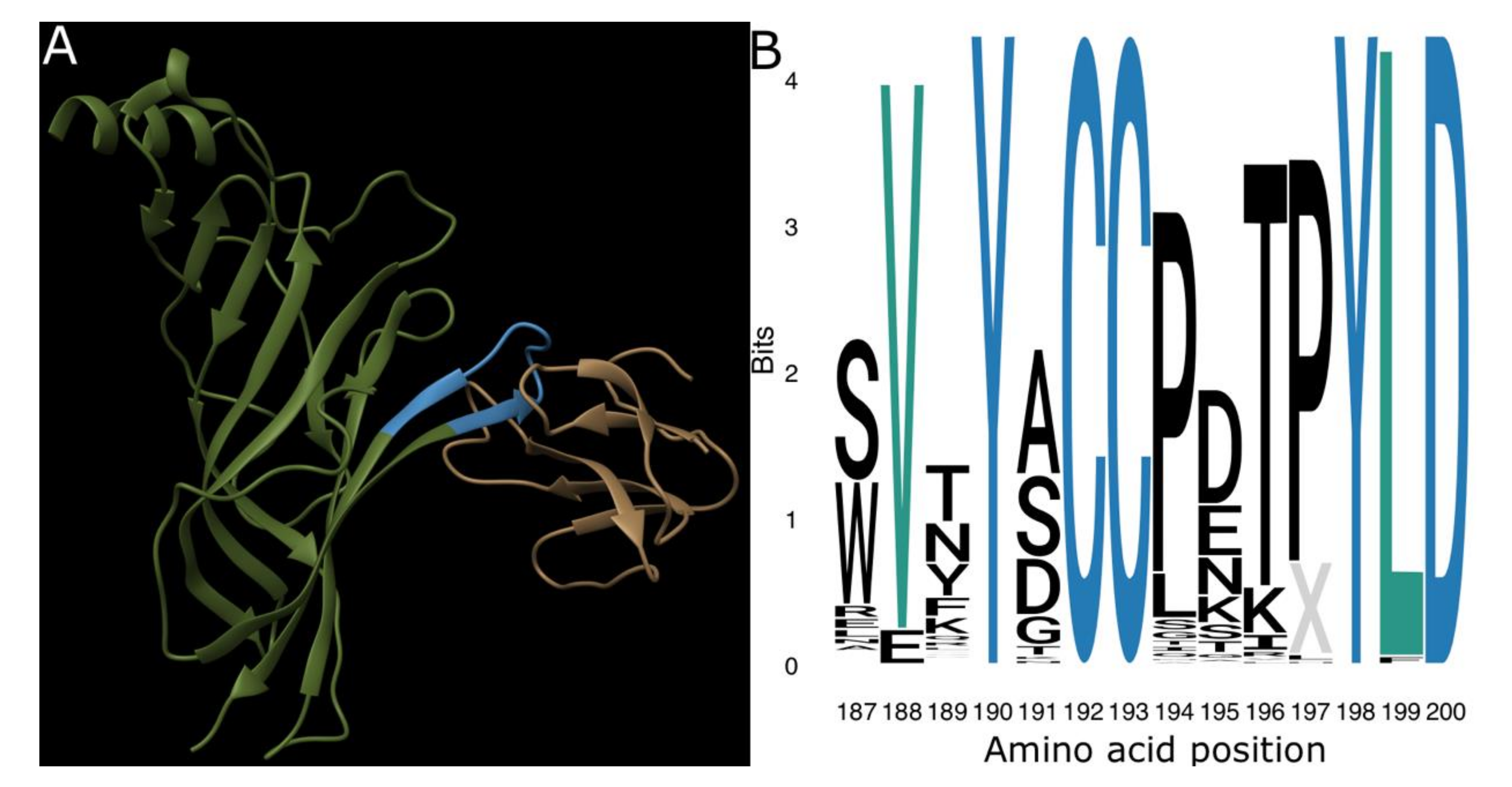
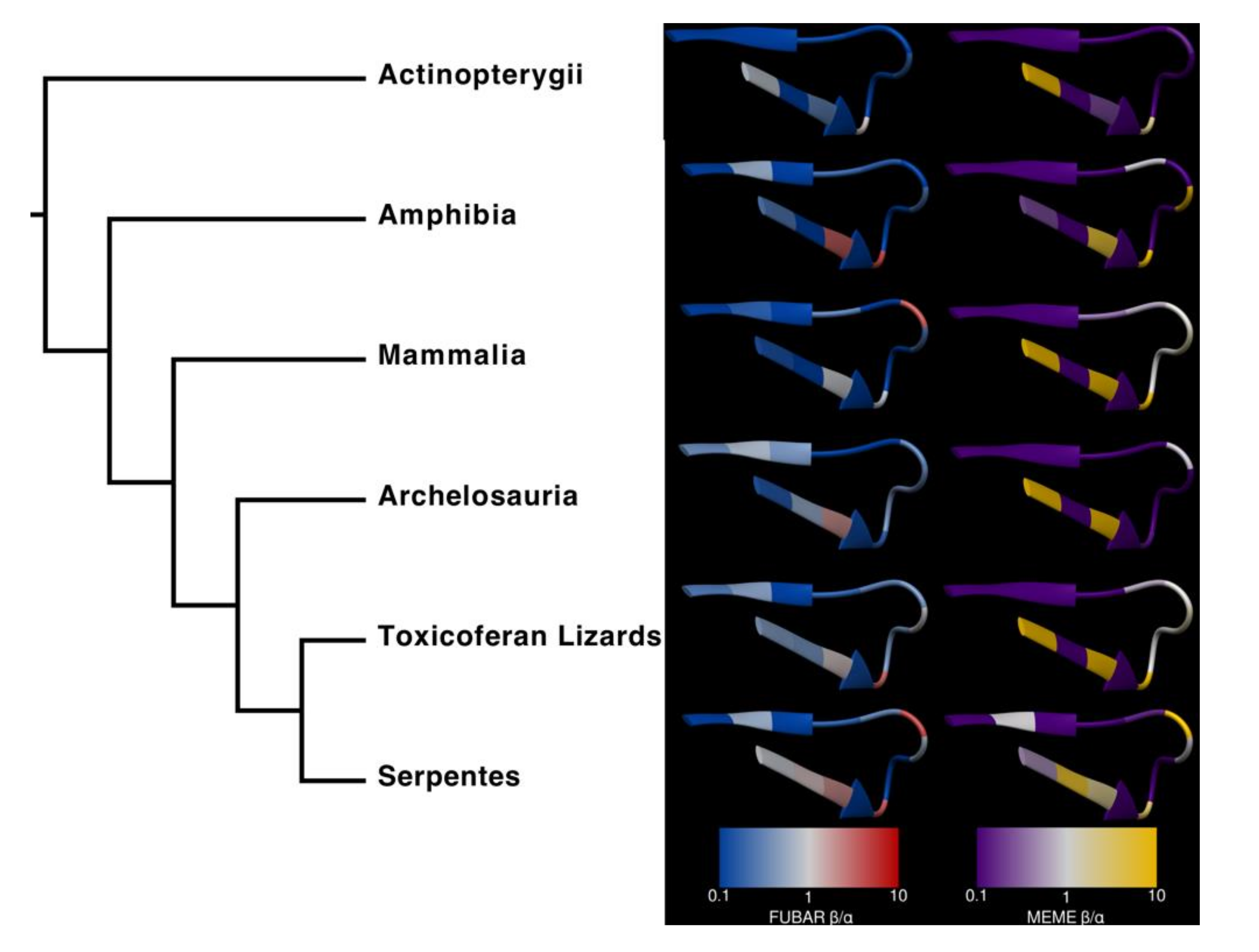
2. Results and Discussion

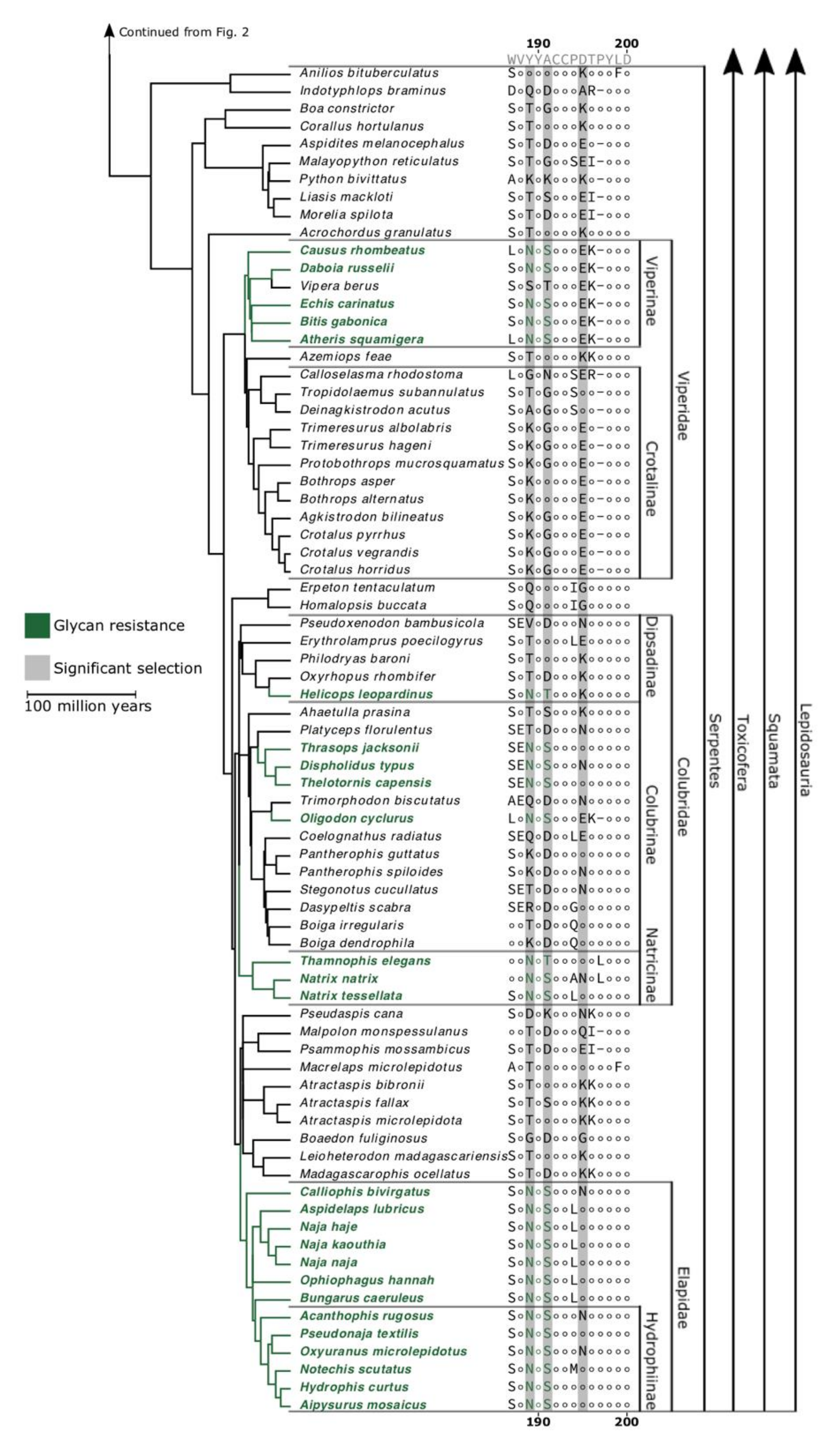
3. Conclusions
4. Materials and Methods
4.1. Ethics Statement
4.2. Tissue Samples
4.3. Amplification and Sequencing of the Ligand-Binding Domain of α-Neurotoxin nAChR
4.4. Analysis of Site-Specific Selection
4.5. Toxicity Assays Using Embryos
4.5.1. Preparation of Venom Stock Solution
4.5.2. Embryo Set-Up
4.5.3. LD50 Assay in Gallus gallus (Domestic Chicken) Embryos
4.5.4. LD50 Assay in Pogona vitticeps (Inland Bearded Dragon) Embryos
4.5.5. LC50 Assay on Gasterosteus aculeatus (Three-Spined Stickleback) and Danio rerio (Zebrafish) Developmental Stages
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.A.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The Toxicogenomic Multiverse: Convergent Recruitment of Proteins Into Animal Venoms. Annu. Rev. Genom. Hum. G 2009, 10, 483–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawkins, R.; Krebs, J.R. Arms races between and within species. Proc. R. Soc. Lond. B Biol. Sci. 1979, 205, 489–511. [Google Scholar] [PubMed]
- Brodie, I.I.I.E.D.; Brodie, J.E.D. Predator-Prey Arms RacesAsymmetrical selection on predators and prey may be reduced when prey are dangerous. BioScience 1999, 49, 557–568. [Google Scholar] [CrossRef]
- Cott, H.B. Adaptive Coloration in Animals; Methuen: London, UK, 1940. [Google Scholar]
- Thompson, J.N. The evolution of species interactions. Science 1999, 284, 2116–2118. [Google Scholar] [CrossRef] [PubMed]
- Vonk, F.J.; Casewell, N.R.; Henkel, C.V.; Heimberg, A.M.; Jansen, H.J.; McCleary, R.J.; Kerkkamp, H.M.; Vos, R.A.; Guerreiro, I.; Calvete, J.J.; et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA 2013, 110, 20651–20656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.C. Looking back on the discovery of alpha-bungarotoxin. J. Biomed. Sci. 1999, 6, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Barber, C.M.; Isbister, G.K.; Hodgson, W.C. Alpha neurotoxins. Toxicon 2013, 66, 47–58. [Google Scholar] [CrossRef]
- Dutertre, S.; Nicke, A.; Tsetlin, V.I. Nicotinic acetylcholine receptor inhibitors derived from snake and snail venoms. Neuropharmacology 2017, 127, 196–223. [Google Scholar] [CrossRef]
- Utkin, Y.; Sunagar, K.; Jackson, T.N.W.; Reeks, T.; Fry, B.G. Three-Finger Toxins (3FTxs); Fry, B.G., Ed.; Oxford University Press: New York, NY, USA, 2015. [Google Scholar]
- Suryamohan, K.; Krishnankutty, S.P.; Guillory, J.; Jevit, M.; Schroder, M.S.; Wu, M.; Kuriakose, B.; Mathew, O.K.; Perumal, R.C.; Koludarov, I.; et al. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat. Genet. 2020, 52, 106–117. [Google Scholar] [CrossRef] [Green Version]
- Bourne, Y.; Talley, T.T.; Hansen, S.B.; Taylor, P.; Marchot, P. Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors. EMBO J. 2005, 24, 1512–1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.C.; Lee, C.Y. Isolation of Neurotoxins from the Venom of Bungarus Multicinctus and Their Modes of Neuromuscular Blocking Action. Arch. Int. Pharm. 1963, 144, 241–257. [Google Scholar]
- Pawlak, J.; Mackessy, S.P.; Sixberry, N.M.; Stura, E.A.; Le Du, M.H.; Menez, R.; Foo, C.S.; Menez, A.; Nirthanan, S.; Kini, R.M. Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity. FASEB J. 2009, 23, 534–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fry, B.G.; Wuster, W.; Ryan Ramjan, S.F.; Jackson, T.; Martelli, P.; Kini, R.M. Analysis of Colubroidea snake venoms by liquid chromatography with mass spectrometry: Evolutionary and toxinological implications. Rapid Commun. Mass Spectrom. 2003, 17, 2047–2062. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Lumsden, N.G.; Wuster, W.; Wickramaratna, J.C.; Hodgson, W.C.; Kini, R.M. Isolation of a neurotoxin (alpha-colubritoxin) from a nonvenomous colubrid: Evidence for early origin of venom in snakes. J. Mol. Evol. 2003, 57, 446–452. [Google Scholar] [CrossRef]
- Chauhan, V.; Thakur, S. The North-South divide in snake bite envenomation in India. J. EmergenciesTraumaShock 2016, 9, 151–154. [Google Scholar] [CrossRef]
- Savidge, J.A. Extinction of an Island Forest Avifauna by an Introduced Snake. Ecology 1987, 68, 660–668. [Google Scholar] [CrossRef]
- Rahman, M.M.; Teng, J.; Worrell, B.T.; Noviello, C.M.; Lee, M.; Karlin, A.; Stowell, M.H.B.; Hibbs, R.E. Structure of the Native Muscle-type Nicotinic Receptor and Inhibition by Snake Venom Toxins. Neuron 2020. [Google Scholar] [CrossRef]
- Edmunds, M. Defence in Animals: A Survey of Anti-Predator Defences; Longman: New York, NY, USA, 1974. [Google Scholar]
- Ujvari, B.; Casewell, N.R.; Sunagar, K.; Arbuckle, K.; Wüster, W.; Lo, N.; O’Meally, D.; Beckmann, C.; King, G.F.; Deplazes, E.; et al. Widespread convergence in toxin resistance by predictable molecular evolution. Proc. Natl. Acad. Sci. USA 2015, 112, 11911–11916. [Google Scholar] [CrossRef] [Green Version]
- Arbuckle, K.; Rodriguez de la Vega, R.C.; Casewell, N.R. Coevolution takes the sting out of it: Evolutionary biology and mechanisms of toxin resistance in animals. Toxicon 2017, 140, 118–131. [Google Scholar] [CrossRef]
- Takacs, Z.; Wilhelmsen, K.C.; Sorota, S. Snake α-Neurotoxin Binding Site on the Egyptian Cobra (Naja haje) Nicotinic Acetylcholine Receptor Is Conserved. Mol. Biol. Evol. 2001, 18, 1800–1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toledo, G.; Hanifin, C.; Geffeney, S.; Brodie, E.D. Convergent Evolution of Tetrodotoxin-Resistant Sodium Channels in Predators and Prey. In Current Topics in Membranes; Robert, J.F., Sergei Yu, N., Eds.; Academic Press: Cambridge, MA, USA, 2016; Volume 78, pp. 87–113. [Google Scholar]
- Venkatesh, B.; Lu, S.Q.; Dandona, N.; See, S.L.; Brenner, S.; Soong, T.W. Genetic basis of tetrodotoxin resistance in pufferfishes. Curr. Biol. 2005, 15, 2069–2072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geffeney, S.L.; Fujimoto, E.; Brodie, E.D., 3rd; Brodie, E.D., Jr.; Ruben, P.C. Evolutionary diversification of TTX-resistant sodium channels in a predator-prey interaction. Nature 2005, 434, 759–763. [Google Scholar] [CrossRef] [PubMed]
- Mellquist, J.L.; Kasturi, L.; Spitalnik, S.L.; Shakin-Eshleman, S.H. The amino acid following an asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry 1998, 37, 6833–6837. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, K.; Marth, J.D. Glycosylation in Cellular Mechanisms of Health and Disease. Cell 2006, 126, 855–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gavel, Y.; von Heijne, G. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: Implications for protein engineering. Protein Eng. 1990, 3, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Takacs, Z.; Wilhelmsen, K.C.; Sorota, S. Cobra (Naja spp.) Nicotinic Acetylcholine Receptor Exhibits Resistance to Erabu Sea Snake ( Laticauda semifasciata ) Short-Chain a-Neurotoxin. J. Mol. Evol. 2004, 58, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Asher, O.; Lupu-Meiri, M.; Jensen, B.S.; Paperna, T.; Fuchs, S.; Oron, Y. Functional characterization of mongoose nicotinic acetylcholine receptor alpha-subunit: Resistance to alpha-bungarotoxin and high sensitivity to acetylcholine. FEBS Lett. 1998, 431, 411–414. [Google Scholar] [CrossRef] [Green Version]
- Kachalsky, S.G.; Jensen, B.S.; Barchan, D.; Fuchs, S. Two subsites in the binding domain of the acetylcholine receptor: An aromatic subsite and a proline subsite. Proc. Natl. Acad. Sci. USA 1995, 92, 10801–10805. [Google Scholar] [CrossRef] [Green Version]
- Wagih, O. ggseqlogo: A versatile R package for drawing sequence logos. Bioinformatics 2017, 33, 3645–3647. [Google Scholar] [CrossRef] [Green Version]
- Barchan, D.; Kachalsky, S.; Neumann, D.; Vogel, Z.; Ovadia, M.; Kochva, E.; Fuchs, S. How the mongoose can fight the snake: The binding site of the mongoose acetylcholine receptor. Proc. Natl. Acad. Sci. USA 1992, 89, 7717–7721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuchs, S.; Barchan, D.; Kachalsky, S.; Neumann, D.; Aladjem, M.; Vogel, Z.; Ovadia, M.; Kochva, E. Molecular evolution of the binding site of the acetylcholine receptor. Ann. N. Y. Acad. Sci. 1993, 681, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Drabeck, D.H.; Dean, A.M.; Jansa, S.A. Why the honey badger don’t care: Convergent evolution of venom-targeted nicotinic acetylcholine receptors in mammals that survive venomous snake bites. Toxicon 2015, 99, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Barchan, D.; Ovadia, M.; Kochva, E.; Fuchs, S. The Binding Site of the Nicotinic Acetylcholine Receptor in Animal Species Resistant to α-Bungarotoxin. Biochemistry 1995, 34, 9172–9176. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.M.; Evans, H.J. Prediction of potential protein-protein interaction sites from amino acid sequence. Identification of a fibrin polymerization site. FEBS Lett. 1996, 385, 81–86. [Google Scholar] [CrossRef] [Green Version]
- Asher, O.; Jensen, B.S.; Lupu-Meiri, M.; Oron, Y.; Fuchs, S. The mongoose acetylcholine receptor alpha-subunit: Analysis of glycosylation and alpha-bungarotoxin binding. FEBS Lett. 1998, 426, 212–216. [Google Scholar] [CrossRef] [Green Version]
- Asher, O.; Lupu-Meiri, M.; Jensen, B.S.; Paperna, T.; Oron, Y.; Fuchs, S. How does the mongoose cope with alpha-bungarotoxin? Analysis of the mongoose muscle AChR alpha-subunit. Ann. N. Y. Acad. Sci. 1998, 841, 97–100. [Google Scholar] [CrossRef]
- Dellisanti, C.D.; Yao, Y.; Stroud, J.C.; Wang, Z.-Z.; Chen, L. Crystal structure of the extracellular domain of nAChR α1 bound to α-bungarotoxin at 1.94 Å resolution. Nat. Neurosci. 2007, 10, 953–962. [Google Scholar] [CrossRef]
- Lerner, H.R.L.; Mindell, D.P. Phylogeny of eagles, Old World vultures, and other Accipitridae based on nuclear and mitochondrial DNA. Mol. Phylogenet. Evol. 2005, 37, 327–346. [Google Scholar] [CrossRef]
- dos Reis, M.; Inoue, J.; Hasegawa, M.; Asher, R.J.; Donoghue, P.C.J.; Yang, Z. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc. R. Soc. B 2012, 279, 3491–3500. [Google Scholar] [CrossRef]
- Prum, R.O.; Berv, J.S.; Dornburg, A.; Field, D.J.; Townsend, J.P.; Lemmon, E.M.; Lemmon, A.R. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 2015, 526, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Alencar, L.R.V.; Quental, T.B.; Grazziotin, F.G.; Alfaro, M.L.; Martins, M.; Venzon, M.; Zaher, H. Diversification in vipers: Phylogenetic relationships, time of divergence and shifts in speciation rates. Mol. Phylogenet. Evol. 2016, 105, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Betancur-R, R.; Wiley, E.O.; Arratia, G.; Acero, A.; Bailly, N.; Miya, M.; Lecointre, G.; Ortí, G. Phylogenetic classification of bony fishes. BMC Evol. Biol. 2017, 17, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef]
- Portillo, F.; Stanley, E.L.; Branch, W.R.; Conradie, W.; Rödel, M.-O.; Penner, J.; Barej, M.F.; Kusamba, C.; Muninga, W.M.; Aristote, M.M.; et al. Evolutionary history of burrowing asps (Lamprophiidae: Atractaspidinae) with emphasis on fang evolution and prey selection. PLoS ONE 2019, 14, e0214889. [Google Scholar] [CrossRef]
- Šmíd, J.; Tolley, K.A. Calibrating the tree of vipers under the fossilized birth-death model. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Zaher, H.; Murphy, R.W.; Arredondo, J.C.; Graboski, R.; Machado-Filho, P.R.; Mahlow, K.; Montingelli, G.G.; Quadros, A.B.; Orlov, N.L.; Wilkinson, M.; et al. Large-scale molecular phylogeny, morphology, divergence-time estimation, and the fossil record of advanced caenophidian snakes (Squamata: Serpentes). PLoS ONE 2019, 14, e0216148. [Google Scholar] [CrossRef] [Green Version]
- Faraggi, D.; Izikson, P.; Reiser, B. Confidence intervals for the 50 per cent response dose. Stat. Med. 2003, 22, 1977–1988. [Google Scholar] [CrossRef]
- Paige, L.; Chapman, P.L.; Butler, R.W. Small sample LD50 confidence intervals using saddlepoint approximations. J. Am. Stat. Assoc. 2011, 106, 334–344. [Google Scholar] [CrossRef]
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-response analysis using R. PLoS ONE 2016, 10, e0146021. [Google Scholar] [CrossRef] [Green Version]
- Harris, R.J.; Zdenek, C.N.; Harrich, D.; Frank, N.; Fry, B.G. An Appetite for Destruction: Detecting Prey-Selective Binding of alpha-Neurotoxins in the Venom of Afro-Asian Elapids. Toxins (Basel) 2020, 12, 205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zdenek, C.N.; Harris, R.J.; Kuruppu, S.; Youngman, N.J.; Dobson, J.S.; Debono, J.; Khan, M.; Smith, I.; Yarski, M.; Harrich, D.; et al. A Taxon-Specific and High-Throughput Method for Measuring Ligand Binding to Nicotinic Acetylcholine Receptors. Toxins (Basel) 2019, 11, 600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dellisanti, C.D.; Yao, Y.; Stroud, J.C.; Wang, Z.Z.; Chen, L. Structural determinants for alpha-neurotoxin sensitivity in muscle nAChR and their implications for the gating mechanism. Channels (Austin) 2007, 1, 234–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dashevsky, D.; Fry, B.G. Ancient Diversification of Three-Finger Toxins in Micrurus Coral Snakes. J. Mol. Evol. 2018, 86, 58–67. [Google Scholar] [CrossRef]
- Aráoz, R.; Molgó, J.; Tandeau de Marsac, N. Neurotoxic cyanobacterial toxins. Toxicon 2010, 56, 813–828. [Google Scholar] [CrossRef]
- Farquhar, C.C. Ecology and Breeding Behavior of the White-Tailed Hawk on the Northern Coastal Prairies of Texas. Master’s Thesis, Texas A&M University, College Station, TX, USA, 1986. [Google Scholar]
- Fitch, H.S.; Bare, R.O. A Field Study of the Red-Tailed Hawk in Eastern Kansas. Trans. Kans. Acad. Sci. (1903-) 1978, 81, 1–13. [Google Scholar] [CrossRef]
- Bent, A.C. Life histories of North American birds of prey. Part 1, Order Falconiformes. In Bulletin United States National Museum no. 167; Smithsonian Institution: Washington, DC, USA, 1937. [Google Scholar]
- Knight, R.L.; Erickson, A.W. High incidence of snakes in the diet of nesting red-tailed hawks. J. Raptor Res. 1976, 10, 108–111. [Google Scholar]
- Chopra, G.; Kumar, T. A study of food and feeding habits of blue peafowl, Pavo Cristatus Linnaeus, 1758 in District Kurukshetra, Haryana (India). Int. J. Res. Stud. Biosci. 2014, 2, 11–16. [Google Scholar]
- Sinclair, I.; Hockey, P.; Tarboton, W. Sasol Birds of Southern Africa; Struik Nature: Cape Town, South Africa, 2012; p. 464. [Google Scholar]
- Pawlak, J.; Mackessy, S.P.; Fry, B.G.; Bhatia, M.; Mourier, G.; Fruchart-Gaillard, C.; Servent, D.; Menez, R.; Stura, E.; Menez, A.; et al. Denmotoxin, a three-finger toxin from the colubrid snake Boiga dendrophila (Mangrove Catsnake) with bird-specific activity. J. Biol. Chem. 2006, 281, 29030–29041. [Google Scholar] [CrossRef] [Green Version]
- Ellemberg, D.; Lewis, T.L.; Liu, C.H.; Maurer, D. Development of spatial and temporal vision during childhood. Vis. Res. 1999, 39, 2325–2333. [Google Scholar] [CrossRef]
- Potier, S.; Lieuvin, M.; Pfaff, M.; Kelber, A. How fast can raptors see? J. Exp. Biol. 2020, 223, jeb209031. [Google Scholar] [CrossRef] [PubMed]
- Oaks, J.R. A time-calibrated species tree of Crocodylia reveals a recent radiation of the true crocodiles. Evolution 2011, 65, 3285–3297. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.Y.; Sanders, K.L.; King, B.; Palci, A. Diversification rates and phenotypic evolution in venomous snakes (Elapidae). Open Sci. 2016, 3, 150277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, S.; Mohandesan, E.; Millar, C.D.; Lambert, D.M. Distance-dependent patterns of molecular divergences in Tuatara mitogenomes. Sci. Rep. 2015, 5, 8703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, R.J.; Zdenek, C.N.; Debono, J.; Harrich, D.; Fry, B.G. Evolutionary Interpretations of Nicotinic Acetylcholine Receptor Targeting Venom Effects by a Clade of Asian Viperidae Snakes. Neurotox Res. 2020. [Google Scholar] [CrossRef]
- Fry, B.G.; Wuster, W.; Kini, R.M.; Brusic, V.; Khan, A.; Venkataraman, D.; Rooney, A.P. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J. Mol. Evol. 2003, 57, 110–129. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Wiens, J.J. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol. Phylogenet. Evol. 2016, 94, 537–547. [Google Scholar] [CrossRef]
- Wallach, V.; Williams, K.L.; Boundy, J. Snakes of the World: A Catalogue of Living and Extinct Species; CRC Press: Boca Raton, FL, USA, 2017; p. 1237. [Google Scholar]
- Carlo, R.E.d.; Leblanc, N.; Brodie, E.D., Jr.; Feldman, C.R. Point-Mutations in Skeletal Muscle Voltage-Gated Sodium Channels Confer Resistance to Tetrodotoxin: But at a Cost? Biophys. J. 2016, 110, 436a–437a. [Google Scholar] [CrossRef]
- Fry, B.G.; Scheib, H.; van der Weerd, L.; Young, B.; McNaughtan, J.; Ramjan, S.F.; Vidal, N.; Poelmann, R.E.; Norman, J.A. Evolution of an arsenal: Structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol. Cell Proteom. 2008, 7, 215–246. [Google Scholar] [CrossRef] [Green Version]
- Pla, D.; Sanz, L.; Whiteley, G.; Wagstaff, S.C.; Harrison, R.A.; Casewell, N.R.; Calvete, J.J. What killed Karl Patterson Schmidt? Combined venom gland transcriptomic, venomic and antivenomic analysis of the South African green tree snake (the boomslang), Dispholidus typus. Biochim. Biophys. Acta (BBA) Gen. Subj. 2017, 1861, 814–823. [Google Scholar] [CrossRef] [Green Version]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- Pond, S.L.K.; Frost, S.D.W.; Muse, S.V. HyPhy: Hypothesis testing using phylogenies. Bioinformatics (Oxford) 2005, 21, 676–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Pond, S.L.K.; Scheffler, K. FUBAR: A Fast, Unconstrained Bayesian AppRoximation for Inferring Selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Pond, S.L.K. Detecting Individual Sites Subject to Episodic Diversifying Selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, Y.Y.; el-Din, M.N.; Mohamed, A.H.; Moustafa, F.A. Effect of cobra venom on the chick embryo. Toxicon 1974, 12, 325. [Google Scholar] [CrossRef]
- Hamburger, V.; Hamilton, H.L. A series of normal stages in the development of the chick embryo. J. Morphol. 1951, 88, 49–92. [Google Scholar] [CrossRef]
| Site | Mutation | Mechanism | Reference |
|---|---|---|---|
| 187 | NXS/T | Steric | [35] |
| R | Steric | [38] | |
| 189 | NXS/T | Steric | [24] |
| 194 | L | Proline | [33] |
| S | Proline | [33] | |
| 197 | H | Proline | [33] |
| Concentration of Naja naja Venom (mg/mL) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.00 | 0.03 | 0.06 | 0.12 | 0.24 | 0.48 | 0.945 | 1.89 | 3.78 | 7.7 | LD50 or LC50 mg/mL | |
| Pogona vitticeps (inland bearded dragon) | 1.87 | ||||||||||
| alive | 5 | - | - | 5 | 5 | 5 | - | - | - | 0 | |
| dead | 0 | - | - | 0 | 0 | 0 | - | - | - | 5 | |
| Gallus gallus (domestic chicken) | 0.340 | ||||||||||
| alive | 5 | - | - | 5 | 5 | 0 | - | - | - | 0 | |
| dead | 0 | - | - | 0 | 0 | 5 | - | - | - | 5 | |
| Gasterosteus aculeatus (three-spined stickleback) | 0.673 | ||||||||||
| alive | 8 | 8 | 8 | 8 | 8 | 8 | 0 | 0 | 0 | 0 | |
| dead | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 8 | 8 | 8 | |
| Danio rerio (zebrafish) | 0.062 | ||||||||||
| alive | 8 | 8 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| dead | 0 | 0 | 3 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.A.; Dashevsky, D.; Kerkkamp, H.; Kordiš, D.; de Bakker, M.A.G.; Wouters, R.; van Thiel, J.; op den Brouw, B.; Vonk, F.J.; Kini, R.M.; et al. Widespread Evolution of Molecular Resistance to Snake Venom α-Neurotoxins in Vertebrates. Toxins 2020, 12, 638. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12100638
Khan MA, Dashevsky D, Kerkkamp H, Kordiš D, de Bakker MAG, Wouters R, van Thiel J, op den Brouw B, Vonk FJ, Kini RM, et al. Widespread Evolution of Molecular Resistance to Snake Venom α-Neurotoxins in Vertebrates. Toxins. 2020; 12(10):638. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12100638
Chicago/Turabian StyleKhan, Muzaffar A., Daniel Dashevsky, Harald Kerkkamp, Dušan Kordiš, Merijn A. G. de Bakker, Roel Wouters, Jory van Thiel, Bianca op den Brouw, Freek J. Vonk, R. Manjunatha Kini, and et al. 2020. "Widespread Evolution of Molecular Resistance to Snake Venom α-Neurotoxins in Vertebrates" Toxins 12, no. 10: 638. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12100638





