Methicillin-Resistant Staphylococcus aureus ST80 Clone: A Systematic Review
Abstract
:1. Introduction
2. Results
2.1. Clonal Complex 80 (CC80)
2.2. Geographical Distribution of MRSA-ST80 in Humans
2.2.1. Asia
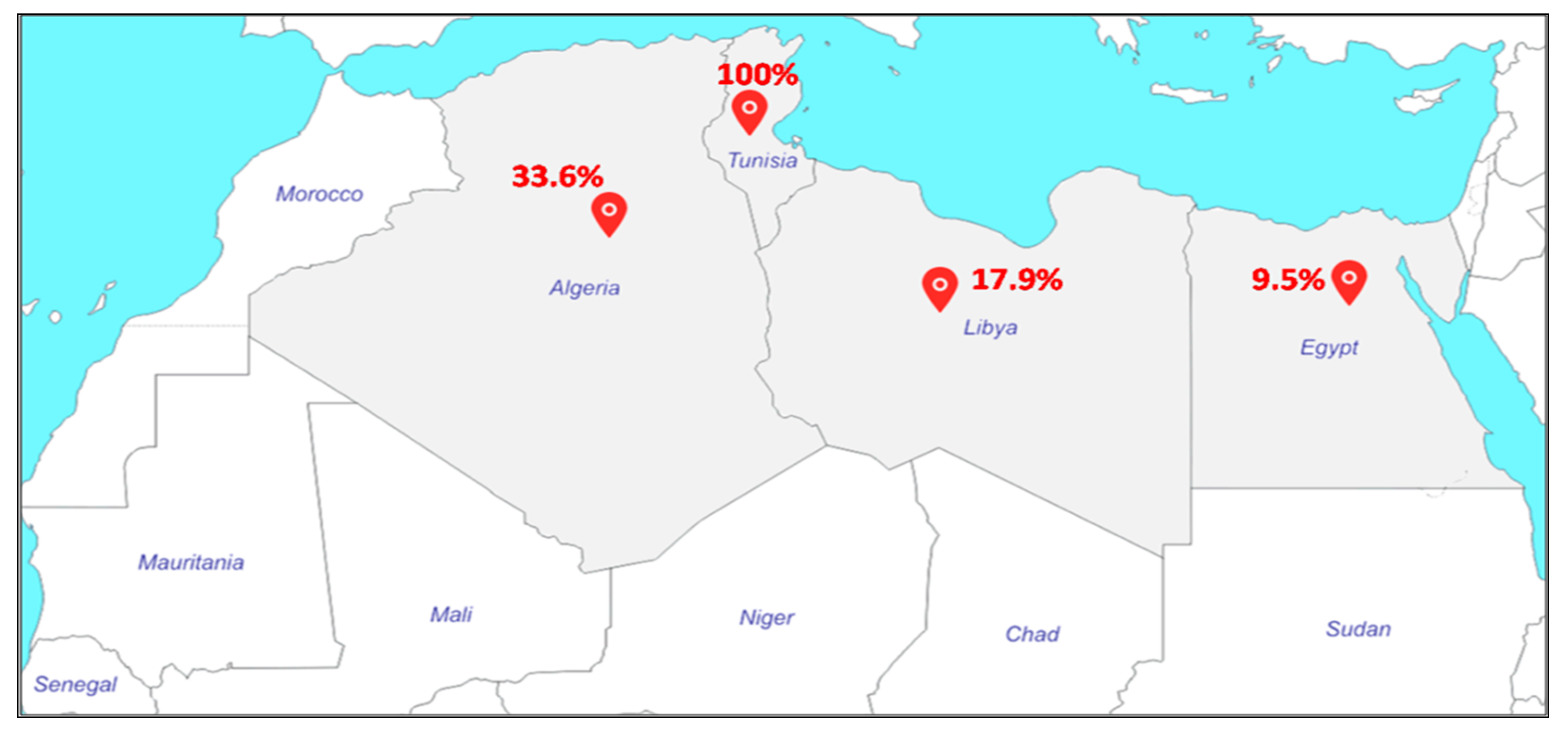
2.2.2. North Africa
2.2.3. Middle East
2.2.4. Europe
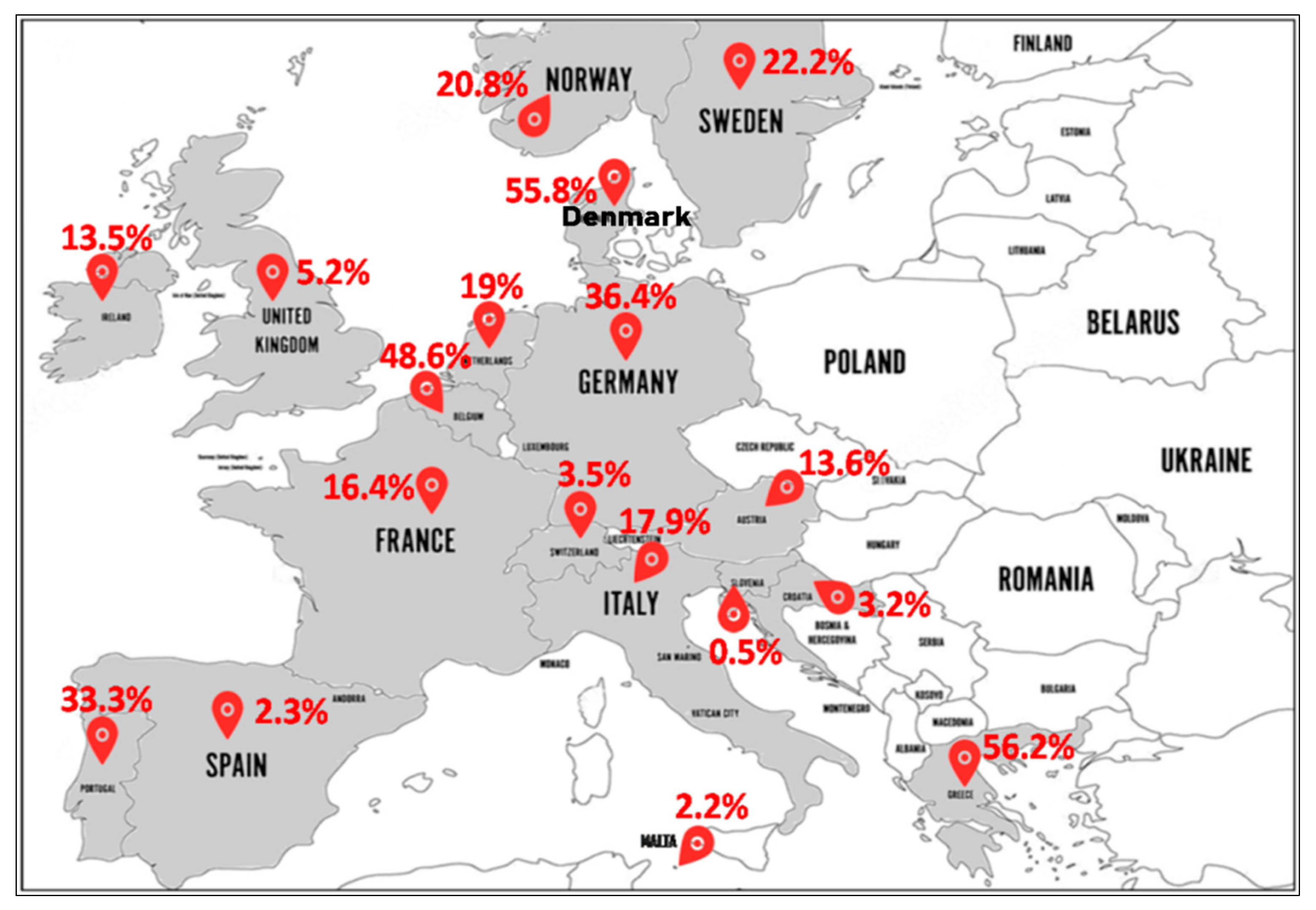
- North Europe (Figure 4): In Norway, MRSA-ST80 frequency was 24.5% (27/110) from 1991 to 2003 [56], 19.4% (13/67) from 1995 to 2003 [57], and 19% (34/179) in 2011 [58]. In Sweden, MRSA-ST80 strains were detected in 33.7% (35/104) [59] and 16.7% (36/216) [60] of all MRSA strains between 2000 and 2005. In Ireland, the reported percentages of MRSA-ST80 among MRSA strains were 8% (2/25) from 1999 to 2005 [61] and 14.2% (27/190) from 2002 to 2011 [62]. In England and Wales, Holmes et al. from 2002 to 2003 reported a low prevalence of PVL-positive S. aureus (1.6%) strains with different genetic backgrounds, although a large number belonged to the ST80 (80% of the PVL+ isolates) that was not geographically related [63]. The emergence of MRSA-ST80 was also noted in London in a collection of MRSA ciprofloxacin-resistant strains between 2000 and 2006 with a prevalence of 2.7% (12/450) [64]. Different percentages of MRSA-ST80 have been reported among MRSA strains in Denmark by many authors as follows: 100% (118/118) from 1997 to 2003 [65], 46.9% (38/81) in 2001 [11], 13.3% (19/143) from 2003 to 2004 [66] and 3.8% (13/341) from 2010 to 2013 [67]. MRSA-ST80 was recognized in 39.2% (206/526) among CA-MRSA strains from 1999 to 2006 [68]. McLaws et al. examined fusidic acid resistance in a collection of 1639 MRSA isolates collected in Denmark between 2003 and 2005 and found that MRSA-ST80 strains accounted for 61.2% (178/291) of the fusidic acid resistant isolates [69].
- South Europe (Figure 4): MRSA-ST80 was reported for the first time in Greece in 2003, which accounted for 9.3% (11/118) of all MRSA strains isolated [21]. Since then, several studies reporting different proportions of MRSA-ST80 among all MRSA isolates in Greece have been published including: 90.0% (18/20) in 2004 [70], 61.7% (428/694) from 2004 to 2005 [71], 100% (27/27) from 2006 to 2007 [72], 33.9% (61/180) from 2003 to 2009 [73], 13.7% (7/51) in 2009 [74], 61.5% (2838/4614) from 2001 to 2012 [75], 78.9% (45/57) from 2010 to. 2011 [76], and 4.8% (19/398) from 2000 to 2015 [77]. Doudoulakakis et al. reported a proportion of 95% (19/20) of selected MRSA isolates among Greek children recovering from pneumonia between 2007 and 2014 [14]. In Croatia, of the 248 MRSA isolates analyzed, 3.2% (8/248) were identified as MRSA-ST80 [17]. In Italy, frequencies of 10% (1/10) [78] and 22.2% (4/18) of MRSA-ST80 among CA-MRSA were reported [79]. In Slovenia, the proportion of MRSA-ST80 recorded was 0.5% (2/385) in a collection of CA-MRSA strains between 2006 and 2013 [80]. In Spain, frequencies of 1.9% (1/53) [81] and 2% (5/246) [82] of MRSA-ST80 among the MRSA PVL+ and CA-MRSA isolates were observed, respectively. Nearly the same percentage of 2.2% (1/45) of MRSA-ST80 among MRSA isolates was reported in Malta [83]. Of the three CA-MRSA strains isolated by Conceição et al., in Portugal, one of these strains was assigned to MRSA-ST80 clone [84].
- West Europe (Figure 4): In Austria, the percentages of MRSA-ST80 among the MRSA PVL+ strains analyzed were respectively 14.9% (14/94) from 2001 to 2006 [85] and 9.7% (3/31) from 2005 to 2010, respectively [86]. In Belgium, data from the reference laboratory for Staphylococci showed that MRSA-ST80 had been detected in 56.3% (9/16) of the MRSA PVL+ strains from 2002 to 2004 [87] and 47.8% (76/159) of the CA-MRSA PVL+ strains from 2005 to 2009 [88]. In Switzerland, the percentage of MRSA-ST80 among MRSA strains was 3.5% (7/200) from 2000 to 2005 [89]. Of the MRSA PVL+ strains described in the Netherlands, the frequency of MRSA-ST80 represented 60% (12/20) from 2000 to 2001 [90], rising to 79.6% (43/54) from 1998 to 2005 [91]. However, the proportion of the MRSA-ST80 clone was 1.1% (2/175) in a collection of MRSA collected from 2002 to 2006 [92] and 2.6% in a collection of 117 MRSA strains isolated from 2003 to 2010 [93]. Varying percentages of MRSA-ST80 were recorded in Germany as follows: 44.6% (33/74) of a low oxacillin-resistant isolates in 2005 [94], 68.4% (80/117) of the MRSA PVL+ isolates from 2005 to 2006 [95], 10.0% (8/80) from 2000 to 2007 [96], 0.3% (10/3207) of MRSA strains between two periods (1 February 2004 to 31 January 2005 and 1 February 2010 to 31 January 2011) [97], and 6.4% (6/94) of MRSA strains from 2012 to 2016 [98]. In a prospective multicenter study of MRSA isolates collected between September 2006 and February 2007 by 23 representative randomly selected French hospital laboratories, MRSA-ST80 strains were detected in 3.8% (4/105) of the MRSA strains isolated from invasive blood cultures [99]. In another French multicenter prospective study conducted in 2008, it was reported that among the 333 MRSA strains isolated, 91 (27.3%) were MRSA-ST80 [100]. Maugat et al. reported a low proportion of MRSA-ST80 of 0.4% (1/238) from 45 private-sector community-based medical laboratories in 2003 [101]. In a study conducted on S. aureus strains isolated from clinically relevant CA-SSTIs in 71 hospitals in November 2006, only one MRSA-ST80 strain (2.9%) was detected from the 34 CA-MRSA strains isolated [102].
2.2.5. Other Parts of the World
2.3. MRSA-ST80 in Extra-Human Niches
2.4. MRSA-ST80 Spread in the Community
2.5. Antibiotic Susceptibility of MRSA-ST80
2.6. Virulence of MRSA-ST80
2.7. MRSA Typing Methods and Techniques
3. Discussion
4. Materials and Methods
4.1. Literature Review
4.2. Data Abstraction
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Peacock, S.J.; Paterson, G.K. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. MRSA virulence and spread. Cell Microbiol. 2012, 14, 1513–1521. [Google Scholar] [CrossRef] [Green Version]
- Harkins, C.P.; Pichon, B.; Doumith, M. Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol. 2017, 18, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dastgheyb, S.S.; Otto, M. Staphylococcal adaptation to diverse physiologic niches: An overview of transcriptomic and phenotypic changes in different biological environments. Future Microbiol. 2015, 10, 1981–1995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.; Wilson, B.; Gould, I.M. Current and future treatment options for community-associated MRSA infection. Expert Opin. Pharmacother. 2018, 19, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Asghar, A.H. Molecular characterization of methicillin-resistant Staphylococcus aureus isolated from tertiary care hospitals. Pak. J. Med. Sci. 2014, 30, 698–702. [Google Scholar] [CrossRef]
- Otter, J.A.; French, G.L. Community-associated meticillin-resistant Staphylococcus aureus: The case for a genotypic definition. J. Hospit. Infect. 2012, 81, 143–148. [Google Scholar] [CrossRef]
- Valle, D.L.; Paclibare, P.A.P.; Cabrera, E.C.; Rivera, W.L. Molecular and phenotypic characterization of methicillin-resistant Staphylococcus aureus isolates from a tertiary hospital in the Philippines. Trop. Med. Health 2016, 44, 3. [Google Scholar] [CrossRef] [Green Version]
- Lakhundi, S.; Zhang, K. Methicillin-resistant Staphylococcus aureus: Molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef] [Green Version]
- Stegger, M.; Wirth, T.; Andersen, P.S.; Skov, R.L.; De Grassi, A.; Simões, P.M.; Tristan, A.; Petersen, A.; Aziz, M.; Kiil, K.; et al. Origin and evolution of european community-acquired methicillin-resistant Staphylococcus aureus. mBio 2014, 5, e01044. [Google Scholar] [CrossRef] [Green Version]
- Faria, N.A.; Oliveira, D.C.; Westh, H.; Monnet, D.L.; Larsen, A.R.; Skov, R.; de Lencastre, H. Epidemiology of emerging methicillin-resistant Staphylococcus aureus (MRSA) in Denmark: A nationwide study in a country with low prevalence of MRSA infection. J. Clin. Microbiol. 2005, 43, 1836–1842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsson, A.-K.; Gustafsson, E.; Johansson, P.J.H.; Odenholt, I.; Petersson, A.C.; Melander, E. Epidemiology of MRSA in southern Sweden: Strong relation to foreign country of origin, health care abroad and foreign travel. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Ehricht, R.; Slickers, P.; Wiese, N.; Jonas, D. Intra-strain variability of methicillin-resistant Staphylococcus aureus strains ST228-MRSA-I and ST5-MRSA-II. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 1383–1390. [Google Scholar] [CrossRef] [PubMed]
- Doudoulakakis, A.G.; Bouras, D.; Drougka, E.; Kazantzi, M.; Michos, A.; Charisiadou, A.; Spiliopoulou, I.; Lebessi, E.; Tsolia, M. Community-associated Staphylococcus aureus pneumonia among Greek children: Epidemiology, molecular characteristics, treatment, and outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1177–1185. [Google Scholar] [CrossRef] [PubMed]
- Abou Shady, H.M.; Bakr, A.E.A.; Hashad, M.E.; Alzohairy, M.A. Staphylococcus aureus nasal carriage among outpatients attending primary health care centers: A comparative study of two cities in Saudi Arabia and Egypt. Braz. J. Infect. Dis. 2015, 19, 68–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alkharsah, K.R.; Rehman, S.; Alkhamis, F.; Alnimr, A.; Diab, A.; Al-Ali, A.K. Comparative and molecular analysis of MRSA isolates from infection sites and carrier colonization sites. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 7. [Google Scholar] [CrossRef] [Green Version]
- Budimir, A.; Deurenberg, R.H.; Bosnjak, Z.; Stobberingh, E.E.; Cetkovic, H.; Kalenic, S. A variant of the Southern German clone of methicillin-resistant Staphylococcus aureus is predominant in Croatia. Clin. Microbiol. Infect. 2010, 16, 1077–1083. [Google Scholar] [CrossRef] [Green Version]
- Edslev, S.M.; Westh, H.; Andersen, P.S.; Skov, R.; Kobayashi, N.; Bartels, M.D.; Vandenesch, F.; Petersen, A.; Worning, P.; Larsen, A.R.; et al. Identification of a PVL-negative SCCmec-IVa sublineage of the methicillin-resistant Staphylococcus aureus CC80 lineage: Understanding the clonal origin of CA-MRSA. Clin. Microbiol. Infect. 2018, 24, 273–278. [Google Scholar] [CrossRef] [Green Version]
- Boswihi, S.S.; Udo, E.E.; Al-Sweih, N. Shifts in the clonal distribution of methicillin-resistant Staphylococcus aureus in Kuwait hospitals: 1992–2010. PLoS ONE 2016, 11, e0162744. [Google Scholar] [CrossRef]
- Udo, E.E.; Sarkhoo, E. The dissemination of ST80-SCCmec-IV community-associated methicillin resistant Staphylococcus aureus clone in Kuwait hospitals. Ann. Clin. Microbiol. Antimicrob. 2010, 9, 31. [Google Scholar] [CrossRef]
- Aires de Sousa, M.; Bartzavali, C.; Spiliopoulou, I.; Sanches, I.S.; Crisóstomo, M.I.; de Lencastre, H. Two international methicillin-resistant Staphylococcus aureus clones endemic in a university hospital in Patras, Greece. J. Clin. Microbiol. 2003, 41, 2027–2032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macedo-Viñas, M.; Conly, J.; Francois, P.; Aschbacher, R.; Blanc, D.S.; Coombs, G.; Daikos, G.; Dhawan, B.; Empel, J.; Etienne, J.; et al. Antibiotic susceptibility and molecular epidemiology of Panton-Valentine leukocidin-positive meticillin-resistant Staphylococcus aureus: An international survey. J. Glob. Antimicrob. Resist. 2014, 2, 43–47. [Google Scholar] [CrossRef] [PubMed]
- El-Mahdy, T.S.; El-Ahmady, M.; Goering, R.V. Molecular characterization of methicillin-resistant Staphylococcus aureus isolated over a 2-year period in a Qatari hospital from multinational patients. Clin. Microbiol. Infect. 2014, 20, 169–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, N.; Ruzan, I.N.; Abd Ghani, M.K.; Hussin, A.; Nawi, S.; Aziz, M.N.; Maning, N.; Eow, V.L.K. Characteristics of community- and hospital-acquired meticillin-resistant Staphylococcus aureus strains carrying SCCmec type IV isolated in Malaysia. J. Med. Microbiol. 2009, 58, 1213–1218. [Google Scholar] [CrossRef]
- Haque, N.; Aung, M.S.; Paul, S.K.; Bari, M.S.; Ahmed, S.; Sarkar, S.R.; Roy, S.; Nasreen, S.A.; Mahmud, M.C.; Hossain, M.A.; et al. Molecular epidemiological characterization of methicillin-susceptible and -resistant Staphylococcus aureus isolated from skin and soft tissue infections in Bangladesh. Microb. Drug Resist. 2019, 25, 241–250. [Google Scholar] [CrossRef]
- Latha, T.; Anil, B.; Manjunatha, H.; Chiranjay, M.; Elsa, D.; Baby, N.; Anice, G. MRSA: The leading pathogen of orthopedic infection in a tertiary care hospital, South India. Afr. Health Sci. 2019, 19, 1393–1401. [Google Scholar] [CrossRef] [Green Version]
- Mendes, R.E.; Mendoza, M.; Banga Singh, K.K.; Castanheira, M.; Bell, J.M.; Turnidge, J.D.; Lin, S.S.F.; Jones, R.N. Regional resistance surveillance program results for 12 Asia-Pacific nations (2011). Antimicrob. Agents Chemother. 2013, 57, 5721–5726. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-J.; Huang, Y.-C. New epidemiology of Staphylococcus aureus infection in Asia. Clin. Microbiol. Infect. 2014, 20, 605–623. [Google Scholar] [CrossRef] [Green Version]
- Ben, N.M.; Merghni, A.; Mastouri, M. Genotyping of Methicillin Resistant Staphylococcus aureus Strains Isolated from Hospitalized Children. Int. J. Pediatr. 2014, 2014, 314316. [Google Scholar]
- Ben Nejma, M.; Mastouri, M.; Bel Hadj Jrad, B.; Nour, M. Characterization of ST80 Panton-Valentine leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus clone in Tunisia. Diagn. Microbiol. Infect. Dis. 2013, 77, 20–24. [Google Scholar] [CrossRef]
- Ahmed, M.O.; Baptiste, K.E.; Daw, M.A.; Elramalli, A.K.; Abouzeed, Y.M.; Petersen, A. Spa typing and identification of pvl genes of meticillin-resistant Staphylococcus aureus isolated from a Libyan hospital in Tripoli. J. Glob. Antimicrob. Resist. 2017, 10, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Antri, K.; Rouzic, N.; Dauwalder, O.; Boubekri, I.; Bes, M.; Lina, G.; Vandenesch, F.; Tazir, M.; Ramdani-Bouguessa, N.; Etienne, J. High prevalence of methicillin-resistant Staphylococcus aureus clone ST80-IV in hospital and community settings in Algiers. Clin. Microbiol. Infect. 2011, 17, 526–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djoudi, F.; Bonura, C.; Benallaoua, S.; Touati, A.; Touati, D.; Aleo, A.; Cala, C.; Fasciana, T.; Mammina, C. Panton-Valentine leukocidin positive sequence type 80 methicillin-resistant Staphylococcus aureus carrying a Staphylococcal cassette chromosome mec type IVc is dominant in neonates and children in an Algiers hospital. New Microbiol. 2013, 36, 49–55. [Google Scholar] [PubMed]
- Alioua, M.A.; Labid, A.; Amoura, K.; Bertine, M.; Gacemi-Kirane, D.; Dekhil, M. Emergence of the European ST80 clone of community-associated methicillin-resistant Staphylococcus aureus as a cause of healthcare-associated infections in Eastern Algeria. Med. Mal. Infect. 2014, 44, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Djahmi, N.; Messad, N.; Nedjai, S.; Moussaoui, A.; Mazouz, D.; Richard, J.-L.; Sotto, A.; Lavigne, J.-P. Molecular epidemiology of Staphylococcus aureus strains isolated from inpatients with infected diabetic foot ulcers in an Algerian University Hospital. Clin. Microbiol. Infect. 2013, 19, E398–E404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bekkhoucha, S.N.; Cady, A.; Gautier, P.; Itim, F.; Donnio, P.-Y. A portrait of Staphylococcus aureus from the other side of the Mediterranean Sea: Molecular characteristics of isolates from Western Algeria. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 553–555. [Google Scholar] [CrossRef]
- Djoudi, F.; Benallaoua, S.; Aleo, A.; Touati, A.; Challal, M.; Bonura, C.; Mammina, C. Descriptive epidemiology of nasal carriage of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus among patients admitted to two healthcare facilities in Algeria. Microb. Drug Resist. 2015, 21, 218–223. [Google Scholar] [CrossRef] [Green Version]
- Oksuz, L.; Dupieux, C.; Tristan, A.; Bes, M.; Etienne, J.; Gurler, N. The high diversity of MRSA clones detected in a university hospital in istanbul. Int. J. Med. Sci. 2013, 10, 1740–1745. [Google Scholar] [CrossRef] [Green Version]
- Udo, E.E.; Al-Lawati, B.A.-H.; Al-Muharmi, Z.; Thukral, S.S. Genotyping of methicillin-resistant Staphylococcus aureus in the Sultan Qaboos University Hospital, Oman reveals the dominance of Panton–Valentine leucocidin-negative ST6-IV/t304 clone. New Microbes New Infect. 2014, 2, 100–105. [Google Scholar] [CrossRef] [Green Version]
- Enany, S.; Yaoita, E.; Yoshida, Y.; Enany, M.; Yamamoto, T. Molecular characterization of Panton-Valentine leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus isolates in Egypt. Microbiol. Res. 2010, 165, 152–162. [Google Scholar] [CrossRef]
- Sonnevend, Á.; Blair, I.; Alkaabi, M.; Jumaa, P.; Al Haj, M.; Ghazawi, A.; Akawi, N.; Jouhar, F.S.; Hamadeh, M.B.; Pál, T. Change in meticillin-resistant Staphylococcus aureus clones at a tertiary care hospital in the United Arab Emirates over a 5-year period. J. Clin. Pathol. 2012, 65, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Laham, N.A.; Mediavilla, J.R.; Chen, L.; Abdelateef, N.; Elamreen, F.A.; Ginocchio, C.C.; Pierard, D.; Becker, K.; Kreiswirth, B.N. MRSA clonal complex 22 strains harboring toxic shock syndrome toxin (TSST-1) are endemic in the primary hospital in Gaza, Palestine. PLoS ONE 2015, 10, e0120008. [Google Scholar] [CrossRef] [Green Version]
- Udo, E.E.; O’Brien, F.G.; Al-Sweih, N.; Noronha, B.; Matthew, B.; Grubb, W.B. Genetic lineages of community-associated methicillin-resistant Staphylococcus aureus in Kuwait hospitals. J. Clin. Microbiol. 2008, 46, 3514–3516. [Google Scholar] [CrossRef] [Green Version]
- Alfouzan, W.; Dhar, R.; Udo, E. Genetic lineages of methicillin-resistant Staphylococcus aureus acquired during admission to an intensive care unit of a general hospital. Med. Princ. Pract. 2017, 26, 113–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Udo, E.E.; Al-Sweih, N. Dominance of community-associated methicillin-resistant Staphylococcus aureus clones in a maternity hospital. PLoS ONE 2017, 12, e0179563. [Google Scholar] [CrossRef] [PubMed]
- Bazzoun, D.A.; Harastani, H.H.; Shehabi, A.A.; Tokajian, S.T. Molecular typing of Staphylococcus aureus collected from a major hospital in Amman, Jordan. J. Infect. Dev. Ctries 2014, 8, 441–447. [Google Scholar] [CrossRef] [Green Version]
- Al-Bakri, A.G.; Al-Hadithi, H.; Kasabri, V.; Othman, G.; Kriegeskorte, A.; Becker, K. The epidemiology and molecular characterization of methicillin-resistant Staphylococci sampled from a healthy Jordanian population. Epidemiol. Infect. 2013, 141, 2384–2391. [Google Scholar] [CrossRef]
- Khalil, W.; Hashwa, F.; Shihabi, A.; Tokajian, S. Methicillin-resistant Staphylococcus aureus ST80-IV clone in children from Jordan. Diagn. Microbiol. Infect. Dis. 2012, 73, 228–230. [Google Scholar] [CrossRef]
- Ohadian Moghadam, S.; Modoodi Yaghooti, M.; Pourramezan, N.; Pourmand, M.R. Molecular characterization and antimicrobial susceptibility of the CA-MRSA isolated from healthcare workers, Tehran, Iran. Microb. Pathog. 2017, 107, 409–412. [Google Scholar] [CrossRef]
- Goudarzi, M.; Navidinia, M.; Beiranvand, E.; Goudarzi, H. Phenotypic and molecular characterization of methicillin-resistant Staphylococcus aureus clones carrying the Panton-Valentine leukocidin genes disseminating in Iranian hospitals. Microb. Drug Resist. 2018, 24, 1543–1551. [Google Scholar] [CrossRef]
- Tokajian, S.T.; Khalil, P.A.; Jabbour, D.; Rizk, M.; Farah, M.J.; Hashwa, F.A.; Araj, G.F. Molecular characterization of Staphylococcus aureus in Lebanon. Epidemiol. Infect. 2010, 138, 707–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harastani, H.H.; Araj, G.F.; Tokajian, S.T. Molecular characteristics of Staphylococcus aureus isolated from a major hospital in Lebanon. Int. J. Infect. Dis. 2014, 19, 33–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monecke, S.; Skakni, L.; Hasan, R.; Ruppelt, A.; Ghazal, S.S.; Hakawi, A.; Slickers, P.; Ehricht, R. Characterisation of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC Microbiol. 2012, 12, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Udo, E.E.; Sarkhoo, E. Genetic analysis of high-level mupirocin resistance in the ST80 clone of community-associated meticillin-resistant Staphylococcus aureus. J. Med. Microbiol. 2010, 59, 193–199. [Google Scholar] [CrossRef] [Green Version]
- Rolo, J.; Miragaia, M.; Turlej-Rogacka, A.; Empel, J.; Bouchami, O.; Faria, N.A.; Tavares, A.; Hryniewicz, W.; Fluit, A.C.; de Lencastre, H.; et al. High genetic diversity among community-associated Staphylococcus aureus in Europe: Results from a multicenter study. PLoS ONE 2012, 7, e34768. [Google Scholar] [CrossRef] [Green Version]
- Fossum, A.E.; Bukholm, G. Increased incidence of methicillin-resistant Staphylococcus aureus ST80, novel ST125 and SCCmecIV in the south-eastern part of Norway during a 12-year period. Clin. Microbiol. Infect. 2006, 12, 627–633. [Google Scholar] [CrossRef] [Green Version]
- Hanssen, A.-M.; Fossum, A.; Mikalsen, J.; Halvorsen, D.S.; Bukholm, G.; Sollid, J.U.E. Dissemination of community-acquired methicillin-resistant Staphylococcus aureus clones in northern Norway: Sequence types 8 and 80 predominate. J. Clin. Microbiol. 2005, 43, 2118–2124. [Google Scholar] [CrossRef] [Green Version]
- Monecke, S.; Aamot, H.V.; Stieber, B.; Ruppelt, A.; Ehricht, R. Characterization of PVL-positive MRSA from Norway. APMIS 2014, 122, 580–584. [Google Scholar] [CrossRef]
- Fang, H.; Hedin, G.; Li, G.; Nord, C.E. Genetic diversity of community-associated methicillin-resistant Staphylococcus aureus in southern Stockholm, 2000–2005. Clin. Microbiol. Infect. 2008, 14, 370–376. [Google Scholar] [CrossRef] [Green Version]
- Petersson, A.C.; Olsson-Liljequist, B.; Miörner, H.; Haeggman, S. Evaluating the usefulness of spa typing, in comparison with pulsed-field gel electrophoresis, for epidemiological typing of methicillin-resistant Staphylococcus aureus in a low-prevalence region in Sweden 2000-2004. Clin. Microbiol. Infect. 2010, 16, 456–462. [Google Scholar] [CrossRef] [Green Version]
- Rossney, A.S.; Shore, A.C.; Morgan, P.M.; Fitzgibbon, M.M.; O’Connell, B.; Coleman, D.C. The emergence and importation of diverse genotypes of methicillin-resistant Staphylococcus aureus (MRSA) harboring the Panton-Valentine leukocidin gene (pvl) reveal that pvl is a poor marker for community-acquired MRSA strains in Ireland. J. Clin. Microbiol. 2007, 45, 2554–2563. [Google Scholar] [CrossRef] [Green Version]
- Shore, A.C.; Tecklenborg, S.C.; Brennan, G.I.; Ehricht, R.; Monecke, S.; Coleman, D.C. Panton-Valentine leukocidin-positive Staphylococcus aureus in Ireland from 2002 to 2011: 21 clones, frequent importation of clones, temporal shifts of predominant methicillin-resistant S. aureus clones, and increasing multiresistance. J. Clin. Microbiol. 2014, 52, 859–870. [Google Scholar] [CrossRef] [Green Version]
- Holmes, A.; Ganner, M.; McGuane, S.; Pitt, T.L.; Cookson, B.D.; Kearns, A.M. Staphylococcus aureus isolates carrying Panton-Valentine leucocidin genes in England and Wales: Frequency, characterization, and association with clinical disease. J. Clin. Microbiol. 2005, 43, 2384–2390. [Google Scholar] [CrossRef] [Green Version]
- Otter, J.A.; French, G.L. The emergence of community-associated methicillin-resistant Staphylococcus aureus at a London teaching hospital, 2000–2006. Clin. Microbiol. Infect. 2008, 14, 670–676. [Google Scholar] [CrossRef] [Green Version]
- Urth, T.; Juul, G.; Skov, R.; Schønheyder, H.C. Spread of a methicillin-resistant Staphylococcus aureus ST80-IV clone in a Danish community. Infect. Control Hosp. Epidemiol. 2005, 26, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Bartels, M.D.; Boye, K.; Rhod Larsen, A.; Skov, R.; Westh, H. Rapid increase of genetically diverse methicillin-resistant Staphylococcus aureus, Copenhagen, Denmark. Emerg. Infect. Dis. 2007, 13, 1533–1540. [Google Scholar] [CrossRef]
- Bartels, M.D.; Larner-Svensson, H.; Meiniche, H.; Kristoffersen, K.; Schonning, K.; Nielsen, J.B.; Rohde, S.M.; Christensen, L.B.; Skibsted, A.W.; Jarlov, J.O.; et al. Monitoring meticillin resistant Staphylococcus aureus and its spread in Copenhagen, Denmark, 2013, through routine whole genome sequencing. Euro Surveill. 2015, 20, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.R.; Stegger, M.; Böcher, S.; Sørum, M.; Monnet, D.L.; Skov, R.L. Emergence and characterization of community-associated methicillin-resistant Staphyloccocus aureus infections in Denmark, 1999 to 2006. J. Clin. Microbiol. 2009, 47, 73–78. [Google Scholar] [CrossRef] [Green Version]
- McLaws, F.B.; Larsen, A.R.; Skov, R.L.; Chopra, I.; O’Neill, A.J. Distribution of fusidic acid resistance determinants in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 55, 1173–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vourli, S.; Perimeni, D.; Makri, A.; Polemis, M.; Voyiatzi, A.; Vatopoulos, A. Community acquired MRSA infections in a paediatric population in Greece. Euro Surveill. 2005, 10, 78–79. [Google Scholar] [CrossRef] [PubMed]
- Chini, V.; Petinaki, E.; Meugnier, H.; Foka, A.; Bes, M.; Etienne, J.; Dimitracopoulos, G.; Spiliopoulou, I. Emergence of a new clone carrying Panton-Valentine leukocidin genes and staphylococcal cassette chromosome mec type V among methicillin-resistant Staphylococcus aureus in Greece. Scand. J. Infect. Dis. 2008, 40, 368–372. [Google Scholar] [CrossRef] [PubMed]
- Vourli, S.; Vagiakou, H.; Ganteris, G.; Orfanidou, M.; Polemis, M.; Vatopoulos, A.; Malamou-Ladas, H. High rates of community-acquired, Panton-Valentine leukocidin (PVL)-positive methicillin-resistant S. aureus (MRSA) infections in adult outpatients in Greece. Euro Surveill. 2009, 14. pii: 19089. [Google Scholar]
- Katopodis, G.D.; Grivea, I.N.; Tsantsaridou, A.J.; Pournaras, S.; Petinaki, E.; Syrogiannopoulos, G.A. Fusidic acid and clindamycin resistance in community-associated, methicillin-resistant Staphylococcus aureus infections in children of Central Greece. BMC Infect. Dis. 2010, 10, 351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hadjihannas, L.; Psichogiou, M.; Empel, J.; Kosmidis, C.; Goukos, D.; Bouzala, J.; Georgopoulos, S.; Malhotra-Kumar, S.; Harbarth, S.; Daikos, G.L.; et al. Molecular characteristics of community-associated methicillin-resistant Staphylococcus aureus colonizing surgical patients in Greece. Diagn. Microbiol. Infect. Dis. 2012, 74, 420–422. [Google Scholar] [CrossRef] [PubMed]
- Drougka, E.; Foka, A.; Liakopoulos, A.; Doudoulakakis, A.; Jelastopulu, E.; Chini, V.; Spiliopoulou, A.; Levidiotou, S.; Panagea, T.; Vogiatzi, A.; et al. A 12-year survey of methicillin-resistant Staphylococcus aureus infections in Greece: ST80-IV epidemic? Clin. Microbiol. Infect. 2014, 20, O796–O803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadimitriou-Olivgeris, M.; Drougka, E.; Fligou, F.; Dodou, V.; Kolonitsiou, F.; Filos, K.S.; Anastassiou, E.D.; Petinaki, E.; Marangos, M.; Spiliopoulou, I. Spread of tst–positive Staphylococcus aureus strains belonging to ST30 clone among patients and healthcare workers in two intensive care units. Toxins 2017, 9, 270. [Google Scholar] [CrossRef] [Green Version]
- Nikolaras, G.P.; Papaparaskevas, J.; Samarkos, M.; Tzouvelekis, L.S.; Psychogiou, M.; Pavlopoulou, I.; Goukos, D.; Polonyfi, K.; Pantazatou, A.; Deliolanis, I.; et al. Changes in the Rates and Population Structure of MRSA from bloodstream infections. A Single Center Experience (2000–2015). J. Glob. Antimicrob. Resist. 2019, 17, 117–122. [Google Scholar] [CrossRef]
- Aschbacher, R.; Pichon, B.; Spoladore, G.; Pagani, E.; Innocenti, P.; Moroder, L.; Ganner, M.; Hill, R.; Pike, R.; Ganthaler, O.; et al. High clonal heterogeneity of Panton-Valentine leukocidin-positive meticillin-resistant Staphylococcus aureus strains from skin and soft-tissue infections in the Province of Bolzano, Northern Italy. Int. J. Antimicrob. Agents 2012, 39, 522–525. [Google Scholar] [CrossRef]
- Sanchini, A.; Campanile, F.; Monaco, M.; Cafiso, V.; Rasigade, J.-P.; Laurent, F.; Etienne, J.; Stefani, S.; Pantosti, A. DNA microarray-based characterisation of Panton-Valentine leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus from Italy. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 1399–1408. [Google Scholar] [CrossRef]
- Dermota, U.; Jurca, T.; Harlander, T.; Košir, M.; Zajc, U.; Golob, M.; Zdovc, I.; Košnik, I.G. Infections caused by community-associated methicillin-resistant Staphylococcus aureus european clone (ST80) in Slovenia between 2006 and 2013. Zdr. Varst 2016, 55, 121–125. [Google Scholar] [CrossRef] [Green Version]
- Cercenado, E.; Cuevas, O.; Marín, M.; Bouza, E.; Trincado, P.; Boquete, T.; Padilla, B.; Vindel, A. Community-acquired methicillin-resistant Staphylococcus aureus in Madrid, Spain: Transcontinental importation and polyclonal emergence of Panton-Valentine leukocidin-positive isolates. Diagn. Microbiol. Infect. Dis. 2008, 61, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Vindel, A.; Trincado, P.; Cuevas, O.; Ballesteros, C.; Bouza, E.; Cercenado, E. Molecular epidemiology of community-associated methicillin-resistant Staphylococcus aureus in Spain: 2004–2012. J. Antimicrob. Chemother. 2014, 69, 2913–2919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scicluna, E.A.; Shore, A.C.; Thürmer, A.; Ehricht, R.; Slickers, P.; Borg, M.A.; Coleman, D.C.; Monecke, S. Characterisation of MRSA from Malta and the description of a Maltese epidemic MRSA strain. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Conceição, T.; Aires-de-Sousa, M.; Pona, N.; Brito, M.J.; Barradas, C.; Coelho, R.; Sardinha, T.; Sancho, L.; de Sousa, G.; do Machado, M.C.; et al. High prevalence of ST121 in community-associated methicillin-susceptible Staphylococcus aureus lineages responsible for skin and soft tissue infections in Portuguese children. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 293–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krziwanek, K.; Luger, C.; Sammer, B.; Stumvoll, S.; Stammler, M.; Metz-Gercek, S.; Mittermayer, H. PVL-positive MRSA in Austria. Eur. J. Clin. Microbiol. Infect. Dis. 2007, 26, 931–935. [Google Scholar] [CrossRef] [PubMed]
- Berktold, M.; Grif, K.; Mäser, M.; Witte, W.; Würzner, R.; Orth-Höller, D. Genetic characterization of Panton-Valentine leukocidin-producing methicillin-resistant Staphylococcus aureus in Western Austria. Wien. Klin. Wochenschr. 2012, 124, 709–715. [Google Scholar] [CrossRef]
- Denis, O.; Deplano, A.; De Beenhouwer, H.; Hallin, M.; Huysmans, G.; Garrino, M.G.; Glupczynski, Y.; Malaviolle, X.; Vergison, A.; Struelens, M.J. Polyclonal emergence and importation of community-acquired methicillin-resistant Staphylococcus aureus strains harbouring Panton-Valentine leucocidin genes in Belgium. J. Antimicrob. Chemother. 2005, 56, 1103–1106. [Google Scholar] [CrossRef]
- Brauner, J.; Hallin, M.; Deplano, A.; De Mendonça, R.; Nonhoff, C.; De Ryck, R.; Roisin, S.; Struelens, M.J.; Denis, O. Community-acquired methicillin-resistant Staphylococcus aureus clones circulating in Belgium from 2005 to 2009: Changing epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 613–620. [Google Scholar] [CrossRef]
- Fenner, L.; Widmer, A.F.; Dangel, M.; Frei, R. Distribution of spa types among meticillin-resistant Staphylococcus aureus isolates during a 6 year period at a low-prevalence University Hospital. J. Med. Microbiol. 2008, 57, 612–616. [Google Scholar] [CrossRef]
- Wannet, W.J.B.; Spalburg, E.; Heck, M.E.O.C.; Pluister, G.N.; Tiemersma, E.; Willems, R.J.L.; Huijsdens, X.W.; de Neeling, A.J.; Etienne, J. Emergence of virulent methicillin-resistant Staphylococcus aureus Strains carrying Panton-Valentine leucocidin genes in The Netherlands. J. Clin. Microbiol. 2005, 43, 3341–3345. [Google Scholar] [CrossRef] [Green Version]
- Stam-Bolink, E.M.; Mithoe, D.; Baas, W.H.; Arends, J.P.; Möller, A.V.M. Spread of a methicillin-resistant Staphylococcus aureus ST80 strain in the community of the northern Netherlands. Eur. J. Clin. Microbiol. Infect. Dis. 2007, 26, 723–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nulens, E.; Stobberingh, E.E.; Smeets, E.; van Dessel, H.; Welling, M.A.; Sebastian, S.; van Tiel, F.H.; Beisser, P.S.; Deurenberg, R.H. Genetic diversity of methicillin-resistant Staphylococcus aureus in a tertiary hospital in The Netherlands between 2002 and 2006. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Hetem, D.J.; Westh, H.; Boye, K.; Jarløv, J.O.; Bonten, M.J.M.; Bootsma, M.C.J. Nosocomial transmission of community-associated methicillin-resistant Staphylococcus aureus in Danish Hospitals. J. Antimicrob. Chemother. 2012, 67, 1775–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witte, W.; Pasemann, B.; Cuny, C. Detection of low-level oxacillin resistance in mecA-positive Staphylococcus aureus. Clin. Microbiol. Infect. 2007, 13, 408–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witte, W.; Strommenger, B.; Cuny, C.; Heuck, D.; Nuebel, U. Methicillin-resistant Staphylococcus aureus containing the Panton-Valentine leucocidin gene in Germany in 2005 and 2006. J. Antimicrob. Chemother. 2007, 60, 1258–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monecke, S.; Jatzwauk, L.; Weber, S.; Slickers, P.; Ehricht, R. DNA microarray-based genotyping of methicillin-resistant Staphylococcus aureus strains from Eastern Saxony. Clin. Microbiol. Infect. 2008, 14, 534–545. [Google Scholar] [CrossRef] [Green Version]
- Schaumburg, F.; Köck, R.; Mellmann, A.; Richter, L.; Hasenberg, F.; Kriegeskorte, A.; Friedrich, A.W.; Gatermann, S.; Peters, G.; von Eiff, C.; et al. Population Dynamics among Methicillin-Resistant Staphylococcus aureus Isolates in Germany during a 6-Year Period. J. Clin. Microbiol. 2012, 50, 3186–3192. [Google Scholar] [CrossRef] [Green Version]
- Klein, S.; Menz, M.-D.; Zanger, P.; Heeg, K.; Nurjadi, D. Increase in the prevalence of Panton-Valentine leukocidin and clonal shift in community-onset methicillin-resistant Staphylococcus aureus causing skin and soft-tissue infections in the Rhine-Neckar Region, Germany, 2012-2016. Int. J. Antimicrob. Agents 2019, 53, 261–267. [Google Scholar] [CrossRef]
- Dauwalder, O.; Lina, G.; Durand, G.; Bes, M.; Meugnier, H.; Jarlier, V.; Coignard, B.; Vandenesch, F.; Etienne, J.; Laurent, F. Epidemiology of invasive methicillin-resistant Staphylococcus aureus clones collected in France in 2006 and 2007. J. Clin. Microbiol. 2008, 46, 3454–3458. [Google Scholar] [CrossRef] [Green Version]
- Robert, J.; Tristan, A.; Cavalié, L.; Decousser, J.-W.; Bes, M.; Etienne, J.; Laurent, F.; ONERBA (Observatoire National de l’Epidémiologie de Résistance Bactérienne aux Antibiotiques). Panton-valentine leukocidin-positive and toxic shock syndrome toxin 1-positive methicillin-resistant Staphylococcus aureus: A French multicenter prospective study in 2008. Antimicrob. Agents Chemother. 2011, 55, 1734–1739. [Google Scholar] [CrossRef]
- Maugat, S.; de Rougemont, A.; Aubry-Damon, H.; Reverdy, M.-E.; Georges, S.; Vandenesch, F.; Etienne, J.; Coignard, B. Methicillin-resistant Staphylococcus aureus among a network of French private-sector community-based-medical laboratories. Med. Mal. Infect. 2009, 39, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Lamy, B.; Laurent, F.; Gallon, O.; Doucet-Populaire, F.; Etienne, J.; Decousser, J.-W.; Collège de Bactériologie Virologie Hygiène (ColBVH) Study Group. Antibacterial resistance, genes encoding toxins and genetic background among Staphylococcus aureus isolated from community-acquired skin and soft tissue infections in France: A national prospective survey. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 1279–1284. [Google Scholar] [CrossRef] [PubMed]
- Fluit, A.C.; Carpaij, N.; Majoor, E.A.M.; Weinstein, R.A.; Aroutcheva, A.; Rice, T.W.; Bonten, M.J.M.; Willems, R.J.L. Comparison of an ST80 MRSA strain from the USA with European ST80 strains. J. Antimicrob. Chemother. 2015, 70, 664–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aung, K.T.; Hsu, L.Y.; Koh, T.H.; Hapuarachchi, H.C.; Chau, M.L.; Gutiérrez, R.A.; Ng, L.C. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in retail food in Singapore. Antimicrob. Resist. Infect. Control 2017, 6, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, M.A.; Parveen, S.; Rahman, M.; Huq, M.; Nabi, A.; Khan, Z.U.M.; Ahmed, N.; Wagenaar, J.A. Occurrence and characterization of methicillin resistant Staphylococcus aureus in processed raw foods and ready-to-eat foods in an urban setting of a developing country. Front. Microbiol. 2019, 10, 503. [Google Scholar] [CrossRef] [PubMed]
- Peeters, L.E.J.; Argudín, M.A.; Azadikhah, S.; Butaye, P. Antimicrobial resistance and population structure of Staphylococcus aureus recovered from pigs farms. Vet. Microbiol. 2015, 180, 151–156. [Google Scholar] [CrossRef]
- Agabou, A.; Ouchenane, Z.; Ngba Essebe, C.; Khemissi, S.; Chehboub, M.T.E.; Chehboub, I.B.; Sotto, A.; Dunyach-Remy, C.; Lavigne, J.-P. Emergence of nasal carriage of ST80 and ST152 PVL+ Staphylococcus aureus isolates from livestock in Algeria. Toxins 2017, 9, 301. [Google Scholar] [CrossRef] [Green Version]
- Mairi, A.; Touati, A.; Pantel, A.; Zenati, K.; Martinez, A.Y.; Dunyach-Remy, C.; Sotto, A.; Lavigne, J.-P. Distribution of toxinogenic methicillin-resistant and methicillin-susceptible Staphylococcus aureus from different ecological niches in Algeria. Toxins 2019, 11, 500. [Google Scholar] [CrossRef] [Green Version]
- Witte, W. Community-acquired methicillin-resistant Staphylococcus aureus: What do we need to know? Clin. Microbiol. Infect. 2009, 15, 17–25. [Google Scholar] [CrossRef] [Green Version]
- Huijsdens, X.W.; van Lier, A.M.C.; van Kregten, E.; Verhoef, L.; van Santen-Verheuvel, M.G.; Spalburg, E.; Wannet, W.J.B. Methicillin-resistant Staphylococcus aureus in Dutch soccer team. Emerg. Infect. Dis. 2006, 12, 1584–1586. [Google Scholar] [CrossRef]
- Drougka, E.; Foka, A.; Koutinas, C.K.; Jelastopulu, E.; Giormezis, N.; Farmaki, O.; Sarrou, S.; Anastassiou, E.D.; Petinaki, E.; Spiliopoulou, I. Interspecies spread of Staphylococcus aureus clones among companion animals and human close contacts in a veterinary teaching hospital. A cross-sectional study in Greece. Prev. Vet. Med. 2016, 126, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Kolonitsiou, F.; Papadimitriou-Olivgeris, M.; Spiliopoulou, A.; Drougka, E.; Jelastopulu, E.; Anastassiou, E.D.; Spiliopoulou, I. Methicillin-resistant Staphylococcus aureus ST80 induce lower cytokine production by monocytes as compared to other Sequence Types. Front. Microbiol. 2018, 9, 3310. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.R.; Böcher, S.; Stegger, M.; Goering, R.; Pallesen, L.V.; Skov, R. Epidemiology of european community-associated methicillin-resistant Staphylococcus aureus clonal complex 80 type IV strains isolated in Denmark from 1993 to 2004. J. Clin. Microbiol. 2008, 46, 62–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Giudice, P.; Bes, M.; Hubiche, T.; Blanc, V.; Roudière, L.; Lina, G.; Vandenesch, F.; Etienne, J. Panton-Valentine leukocidin-positive Staphylococcus aureus strains are associated with follicular skin infections. Dermatology 2011, 222, 167–170. [Google Scholar] [CrossRef]
- Harastani, H.H.; Tokajian, S.T. Community-associated methicillin-resistant Staphylococcus aureus clonal complex 80 type IV (CC80-MRSA-IV) isolated from the Middle East: A heterogeneous expanding clonal lineage. PLoS ONE 2014, 9, e103715. [Google Scholar] [CrossRef] [Green Version]
- Dunyach-Remy, C.; Ngba Essebe, C.; Sotto, A.; Lavigne, J.-P. Staphylococcus aureus toxins and diabetic foot ulcers: Role in pathogenesis and interest in diagnosis. Toxins 2016, 8, 209. [Google Scholar] [CrossRef] [Green Version]
- Courjon, J.; Munro, P.; Benito, Y.; Visvikis, O.; Bouchiat, C.; Boyer, L.; Doye, A.; Lepidi, H.; Ghigo, E.; Lavigne, J.-P.; et al. EDIN-B promotes the translocation of Staphylococcus aureus to the bloodstream in the course of pneumonia. Toxins 2015, 7, 4131–4142. [Google Scholar] [CrossRef] [Green Version]
- Gillet, Y.; Dumitrescu, O.; Tristan, A.; Dauwalder, O.; Jahouvey, E.; Floret, D.; Vandenesch, F.; Etienne, J.; Lina, G. Pragmatic management of Panton-Valentine leukocidin-associated staphylococcal diseases. Int. J. Antimicrob. Agents 2011, 38, 457–464. [Google Scholar] [CrossRef] [Green Version]
- Zingg, W.; Holmes, A.; Dettenkofer, M.; Goetting, T.; Secci, F.; Clack, L.; Allegranzi, B.; Magiorakos, A.P.; Pittet, D. Hospital organisation, management, and structure for prevention of health-care-associated infection: A systematic review and expert consensus. Lancet Infect. Dis. 2015, 15, 212–224. [Google Scholar] [CrossRef]
- Al-Obaidi, M.M.J.; Suhaili, Z.; MohdDesa, M.N. Genotyping Approaches for Identification and Characterization of Staphylococcus aureus; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef] [Green Version]
- Boye, K.; Bartels, M.D.; Andersen, I.S.; Møller, J.A.; Westh, H. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I-V. Clin. Microbiol. Infect. 2007, 13, 725–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Chen, D.; Peters, B.M.; Li, L.; Li, B.; Xu, Z.; Shirliff, M.E. Staphylococcal chromosomal cassettes mec (SCCmec): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb. Pathog. 2016, 101, 56–67. [Google Scholar] [CrossRef]
- Rachman, A.R.A.; Suhaili, Z.; MohdDesa, M.N. The evolution and dissemination of methicillin resistance determinant in Staphylococcus aureus. In The Rise of Virulence and Antibiotic Resistance in Staphylococcus aureus; InTech: London, UK, 2017. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, M.; Hogan, P.G.; Satola, S.W.; Crispell, E.; Wylie, T.; Gao, H.; Sodergren, E.; Weinstock, G.M.; Burnham, C.-A.D.; Fritz, S.A. Discriminatory indices of typing methods for epidemiologic analysis of contemporary Staphylococcus aureus strains. Medicine 2015, 94, e1534. [Google Scholar] [CrossRef] [PubMed]
- Malachowa, N.; Sabat, A.; Gniadkowski, M.; Krzyszton-Russjan, J.; Empel, J.; Miedzobrodzki, J.; Kosowska-Shick, K.; Appelbaum, P.C.; Hryniewicz, W. Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J. Clin. Microbiol. 2005, 43, 3095–3100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alkharsah, K.R.; Rehman, S.; Alnimr, A.; Diab, A.; Hawwari, A.; Tokajian, S. Molecular typing of MRSA isolates by spa and PFGE. J. King Saud Univ. Sci. 2019, 31, 999–1004. [Google Scholar] [CrossRef]
- Enright, M.C.; Day, N.P.J.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef] [Green Version]
- Urwin, R.; Maiden, M.C.J. Multi-locus sequence typing: A tool for global epidemiology. Trends Microbiol. 2003, 11, 479–487. [Google Scholar] [CrossRef]
- Boers, S.A.; van der Reijden, W.A.; Jansen, R. High-throughput multilocus sequence typing: Bringing molecular typing to the next level. PLoS ONE 2012, 7, e39630. [Google Scholar] [CrossRef] [Green Version]
- Hallin, M.; Deplano, A.; Denis, O.; De Mendonça, R.; De Ryck, R.; Struelens, M.J. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J. Clin. Microbiol. 2007, 45, 127–133. [Google Scholar] [CrossRef] [Green Version]
- Bumgarner, R. DNA microarrays: Types, applications and their future. Curr. Protoc. Mol. Biol. 2013, 22, 23288464. [Google Scholar]
- Miao, J.; Chen, L.; Wang, J.; Wang, W.; Chen, D.; Li, L.; Li, B.; Deng, Y.; Xu, Z. Current methodologies on genotyping for nosocomial pathogen methicillin-resistant Staphylococcus aureus (MRSA). Microb. Pathog. 2017, 107, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Zhao, P.; Liu, H.; Yu, X.; Qin, Y.; Su, Z.; Wang, S.; Xu, H.; Chen, J. Whole-genome sequencing for the investigation of a hospital outbreak of MRSA in China. PLoS ONE 2016, 11, e0149844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quainoo, S.; Coolen, J.P.M.; van Hijum, S.A.F.T.; Huynen, M.A.; Melchers, W.J.G.; van Schaik, W.; Wertheim, H.F.L. Whole-genome sequencing of bacterial pathogens: The future of nosocomial outbreak analysis. Clin. Microbiol. Rev. 2017, 30, 1015–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarze, K.; Buchanan, J.; Taylor, J.C.; Wordsworth, S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet. Med. 2018, 20, 1122–1130. [Google Scholar] [CrossRef] [Green Version]
- Rolain, J.-M.; Abat, C.; Brouqui, P.; Raoult, D. Worldwide decrease in methicillin-resistant Staphylococcus aureus: Do we understand something? Clin. Microbiol. Infect. 2015, 21, 515–517. [Google Scholar] [CrossRef] [Green Version]
- Walter, J.; Noll, I.; Feig, M.; Weiss, B.; Claus, H.; Werner, G.; Eckmanns, T.; Hermes, J.; Abu Sin, M. Decline in the proportion of methicillin resistance among Staphylococcus aureus isolates from non-invasive samples and in outpatient settings, and changes in the co-resistance profiles: An analysis of data collected within the Antimicrobial Resistance Surveillance Network, Germany 2010 to 2015. BMC Infect. Dis. 2017, 17, 169. [Google Scholar]
- Lee, A.S.; de Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Primers 2018, 4, 18033. [Google Scholar] [CrossRef]
- Köck, R.; Becker, K.; Cookson, B.; van Gemert-Pijnen, J.E.; Harbarth, S.; Kluytmans, J.; Mielke, M.; Peters, G.; Skov, R.L.; Struelens, M.J.; et al. Methicillin-resistant Staphylococcus aureus (MRSA): Burden of disease and control challenges in Europe. Euro Surveill. 2010, 15, 19688. [Google Scholar] [CrossRef]
- de Lencastre, H.; Severina, E.P.; Milch, H.; Thege, M.K.; Tomasz, A. Wide geographic distribution of a unique methicillin-resistant Staphylococcus aureus clone in Hungarian hospitals. Clin. Microbiol. Infect. 1997, 3, 289–296. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, D.C.; Crisóstomo, I.; Santos-Sanches, I.; Major, P.; Alves, C.R.; Aires-de-Sousa, M.; Thege, M.K.; de Lencastre, H. Comparison of DNA sequencing of the protein A gene polymorphic region with other molecular typing techniques for typing two epidemiologically diverse collections of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2001, 39, 574–580. [Google Scholar] [CrossRef] [Green Version]
- Conceição, T.; Aires-de-Sousa, M.; Füzi, M.; Tóth, A.; Pászti, J.; Ungvári, E.; van Leeuwen, W.B.; van Belkum, A.; Grundmann, H.; de Lencastre, H. Replacement of methicillin-resistant Staphylococcus aureus clones in Hungary over time: A 10-year surveillance study. Clin. Microbiol. Infect. 2007, 13, 971–979. [Google Scholar] [CrossRef] [Green Version]
- Asanin, J.; Misic, D.; Aksentijevic, K.; Tambur, Z.; Rakonjac, B.; Kovacevic, I.; Spergser, J.; Loncaric, I. Genetic profiling and comparison of human and animal methicillin-resistant Staphylococcus aureus (MRSA) isolates from Serbia. Antibiotics 2019, 8, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monecke, S.; Gavier-Widén, D.; Hotzel, H.; Peters, M.; Guenther, S.; Lazaris, A.; Loncaric, I.; Müller, E.; Reissig, A.; Ruppelt-Lorz, A.; et al. Diversity of Staphylococcus aureus isolates in european wildlife. PLoS ONE 2016, 11, e0168433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botelho, A.M.N.; Cerqueira, E.; Costa, M.O.; Moustafa, A.M.; Beltrame, C.O.; Ferreira, F.A.; Côrtes, M.F.; Costa, B.S.S.; Silva, D.N.S.; Bandeira, P.T.; et al. Local diversification of methicillin- resistant Staphylococcus aureus ST239 in South America after its rapid worldwide dissemination. Front. Microbiol. 2019, 10, 82. [Google Scholar] [CrossRef]
- Goudarzi, M.; Goudarzi, H.; Sá Figueiredo, A.M.; Udo, E.E.; Fazeli, M.; Asadzadeh, M.; Seyedjavadi, S.S. Molecular characterization of methicillin resistant Staphylococcus aureus strains isolated from intensive care units in Iran: ST22-SCCmec IV/t790 emerges as the major clone. PLoS ONE 2016, 11, e0155529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goudarzi, M.; Eslami, G.; Rezaee, R.; Heidary, M.; Khoshnood, S.; Sajadi Nia, R. Clonal dissemination of Staphylococcus aureus isolates causing nosocomial infections, Tehran, Iran. Iran. J. Basic Med. Sci. 2019, 22, 238–245. [Google Scholar] [PubMed]
- Goudarzi, M.; Kobayashi, N.; Hashemi, A.; Fazeli, M.; Navidinia, M. Genetic variability of methicillin resistant Staphylococcus aureus strains isolated from burns patients. Osong Public Health Res. Perspect. 2019, 10, 170–176. [Google Scholar] [CrossRef]
- Çakıcı, N.; Akçalı, A.; Demirel Zorba, N.N. Antibiotic resistance pattern and spa types of Staphylococcus aureus strains isolated from food business and hospital kitchen employees in Çanakkale, Turkey. Turk. J. Med. Sci. 2019, 49, 675–682. [Google Scholar] [CrossRef]
- Chang, Q.; Abuelaish, I.; Biber, A.; Jaber, H.; Callendrello, A.; Andam, C.P.; Regev-Yochay, G.; Hanage, W.P.; On Behalf of the PICR Study Group. Genomic epidemiology of meticillin-resistant Staphylococcus aureus ST22 widespread in communities of the Gaza Strip, 2009. Euro Surveill. 2018, 23, 363. [Google Scholar] [CrossRef]
- Mobasherizadeh, S.; Shojaei, H.; Azadi, D.; Havaei, S.A.; Rostami, S. Molecular characterization and genotyping of methicillin-resistant Staphylococcus aureus in nasal carriage of healthy Iranian children. J. Med. Microbiol. 2019, 68, 374–378. [Google Scholar] [CrossRef]
- Sobhanipoor, M.H.; Ahmadrajabi, R.; Karmostaji, A.; Saffari, F. Molecular characterization of nasal methicillin resistant Staphylococcus aureus isolates from workers of an automaker company in southeast Iran. APMIS 2017, 125, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Udo, E.E.; Boswihi, S.S.; Al-Sweih, N. High prevalence of toxic shock syndrome toxin-producing epidemic methicillin-resistant Staphylococcus aureus 15 (EMRSA-15) strains in Kuwait hospitals. New Microbes New Infect. 2016, 12, 24–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goudarzi, M.; Fazeli, M.; Pouriran, R.; Eslami, G. Genotype distribution of Panton-Valentine leukocidin (PVL)-positive Staphylococcus aureus strains isolated from wound-related infections: A three-year multi-center study in Tehran, Iran. Jpn. J. Infect. Dis. 2019, 72, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Conceição, T.; de Lencastre, H.; Aires-de-Sousa, M. Carriage of Staphylococcus aureus among Portuguese nursing students: A longitudinal cohort study over four years of education. PLoS ONE 2017, 12, e0188855. [Google Scholar] [CrossRef] [Green Version]
- Groves, M.D.; Crouch, B.; Coombs, G.W.; Jordan, D.; Pang, S.; Barton, M.D.; Giffard, P.; Abraham, S.; Trott, D.J. Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolated from Australian veterinarians. PLoS ONE 2016, 11, e0146034. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, A.C.; Belas, A.; Marques, C.; Cruz, L.; Gama, L.T.; Pomba, C. Risk Factors for nasal colonization by methicillin-resistant staphylococci in healthy humans in professional daily contact with companion animals in Portugal. Microb. Drug Resist. 2018, 24, 434–446. [Google Scholar] [CrossRef]
- Rodríguez-Lázaro, D.; Oniciuc, E.-A.; García, P.G.; Gallego, D.; Fernández-Natal, I.; Dominguez-Gil, M.; Eiros-Bouza, J.M.; Wagner, M.; Nicolau, A.I.; Hernández, M. Detection and characterization of Staphylococcus aureus and methicillin-resistant S. aureus in foods confiscated in EU borders. Front. Microbiol. 2017, 8, 1344. [Google Scholar] [CrossRef]
- Tosas Auguet, O.; Stabler, R.A.; Betley, J.; Preston, M.D.; Dhaliwal, M.; Gaunt, M.; Ioannou, A.; Desai, N.; Karadag, T.; Batra, R.; et al. Frequent undetected ward-based methicillin-resistant Staphylococcus aureus transmission linked to patient sharing between hospitals. Clin. Infect. Dis. 2018, 66, 840–848. [Google Scholar] [CrossRef] [Green Version]
- Worthing, K.A.; Abraham, S.; Pang, S.; Coombs, G.W.; Saputra, S.; Jordan, D.; Wong, H.S.; Abraham, R.J.; Trott, D.J.; Norris, J.M. Molecular characterization of methicillin-resistant Staphylococcus aureus isolated from Australian animals and veterinarians. Microb. Drug Resist. 2018, 24, 203–212. [Google Scholar]
- Gostev, V.; Kruglov, A.; Kalinogorskaya, O.; Dmitrenko, O.; Khokhlova, O.; Yamamoto, T.; Lobzin, Y.; Ryabchenko, I.; Sidorenko, S. Molecular epidemiology and antibiotic resistance of methicillin-resistant Staphylococcus aureus circulating in the Russian Federation. Infect. Genet. Evol. 2017, 53, 189–194. [Google Scholar] [CrossRef]
- Htun, H.L.; Kyaw, W.M.; de Sessions, P.F.; Low, L.; Hibberd, M.L.; Chow, A.; Leo, Y.S. Methicillin-resistant Staphylococcus aureus colonisation: Epidemiological and molecular characteristics in an acute-care tertiary hospital in Singapore. Epidemiol. Infect. 2018, 146, 1785–1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, N.; Yang, J.; Duan, N.; Lu, B.; Wang, L. Community-associated Staphylococcus aureus PVL+ ST22 predominates in skin and soft tissue infections in Beijing, China. Infect. Drug Resist. 2019, 12, 2495–2503. [Google Scholar] [CrossRef] [PubMed] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mairi, A.; Touati, A.; Lavigne, J.-P. Methicillin-Resistant Staphylococcus aureus ST80 Clone: A Systematic Review. Toxins 2020, 12, 119. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12020119
Mairi A, Touati A, Lavigne J-P. Methicillin-Resistant Staphylococcus aureus ST80 Clone: A Systematic Review. Toxins. 2020; 12(2):119. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12020119
Chicago/Turabian StyleMairi, Assia, Abdelaziz Touati, and Jean-Philippe Lavigne. 2020. "Methicillin-Resistant Staphylococcus aureus ST80 Clone: A Systematic Review" Toxins 12, no. 2: 119. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12020119




