Prevalence and Genetic Diversity of Staphylococcal Enterotoxin (-Like) Genes sey, selw, selx, selz, sel26 and sel27 in Community-Acquired Methicillin-Resistant Staphylococcus aureus
Abstract
:1. Introduction
2. Results
2.1. The Prevalence of sey, selw, selx, selz, sel26, and sel27
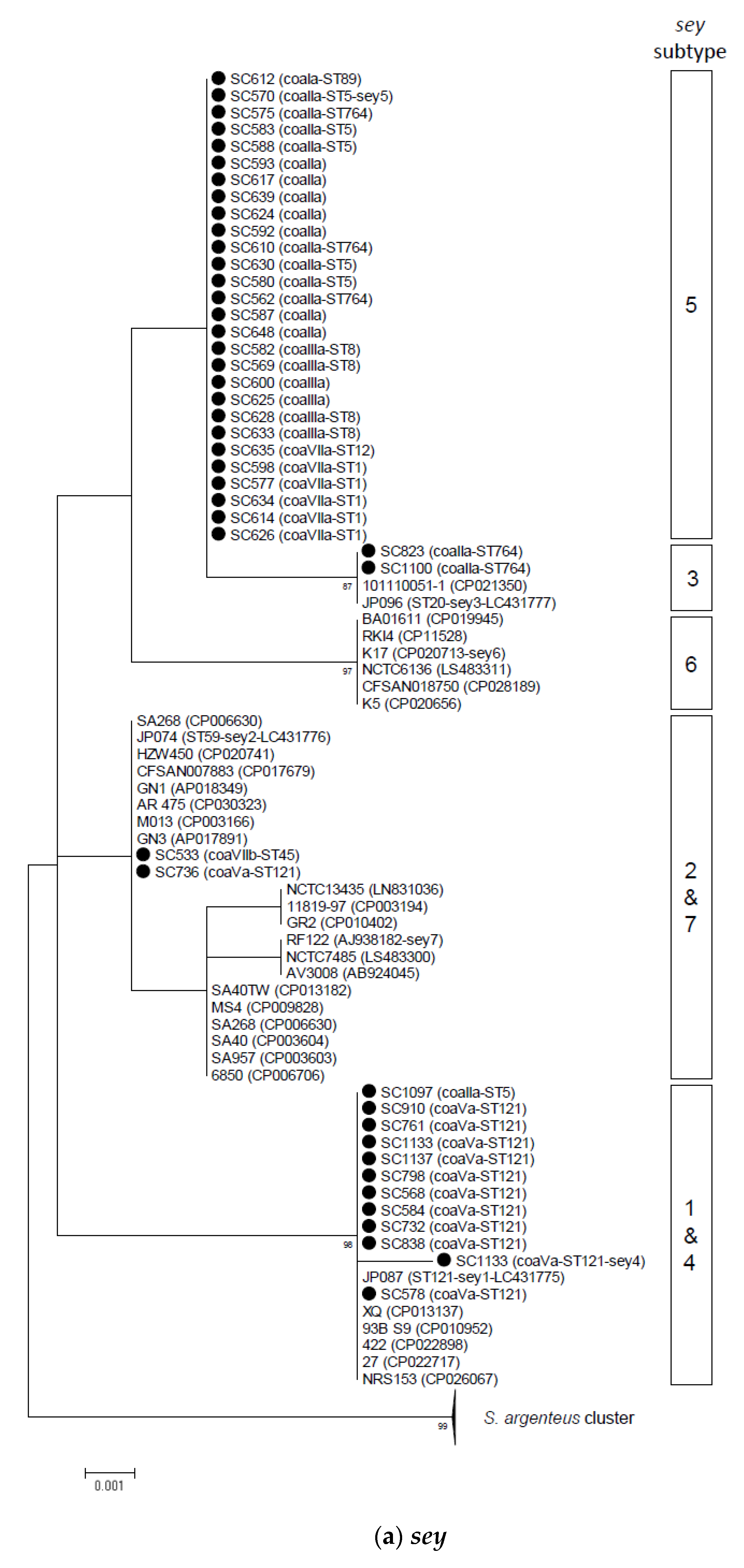
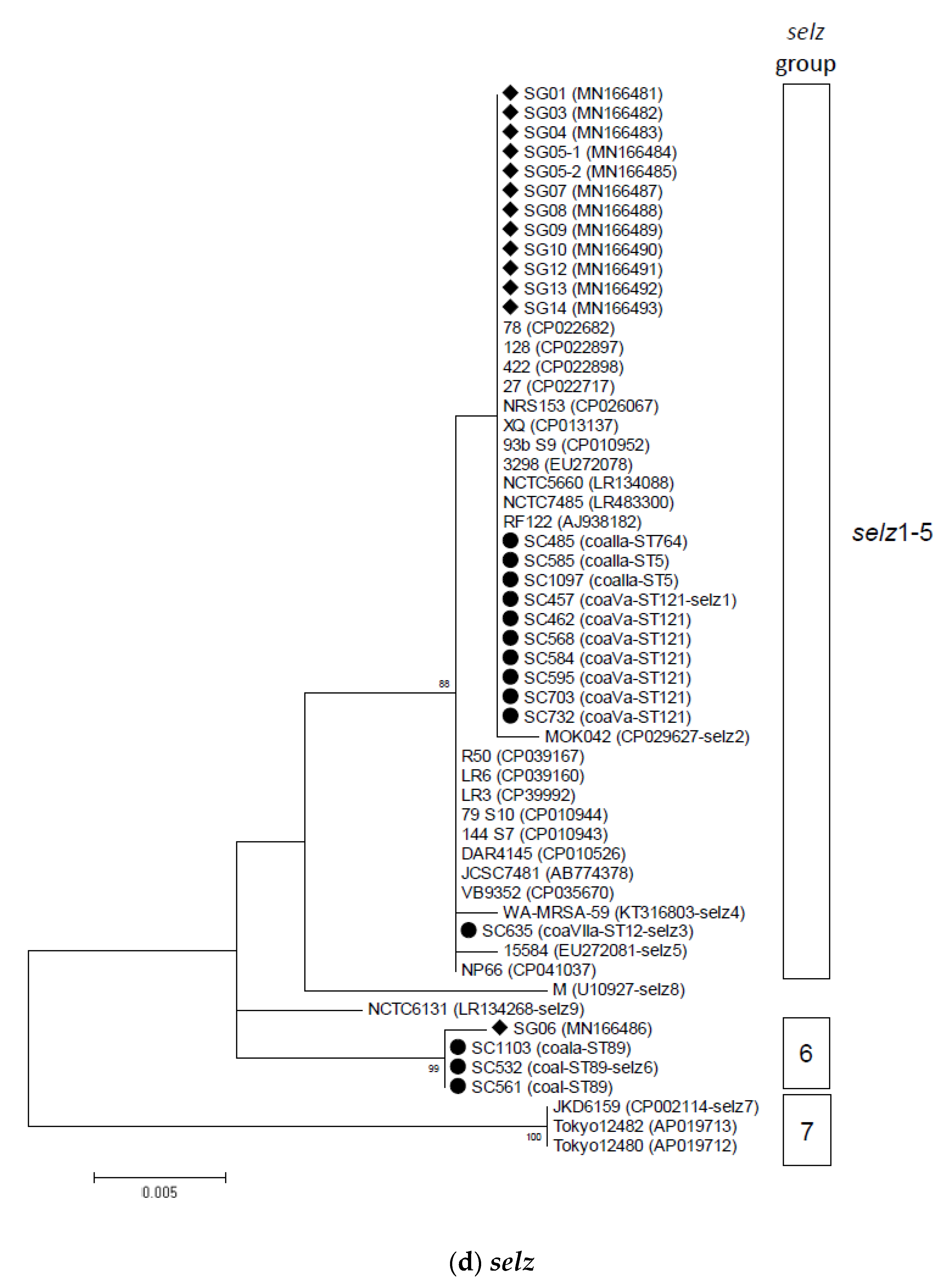
2.2. Phylogenetic and Sequence Analysis of sey, selw, selx, and selz
3. Discussion
4. Materials and Methods
4.1. CA-MRSA Isolates
4.2. Genetic Analysis of S. aureus (CA-MRSA) Isolates
4.3. GenBank Accession Numbers
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- David, M.Z.; Daum, R.S. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010, 23, 616–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spaulding, A.R.; Salgado-Pabón, W.; Kohler, P.L.; Horswill, A.R.; Leung, D.Y.; Schlievert, P.M. Staphylococcal and streptococcal superantigen exotoxins. Clin. Microbiol. Rev. 2013, 26, 422–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, G.J.; Tuffs, S.W.; Wee, B.A.; Seo, K.S.; Park, N.; Connelley, T.; Guinane, C.M.; Morrison, W.I.; Fitzgerald, J.R. Bovine Staphylococcus aureus Superantigens Stimulate the Entire T Cell Repertoire of Cattle. Infect Immun. 2018, 86, e00505–e00518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.F.; Yang, X.Y.; Zhang, J.; Qin, X.; Huang, X.; Cui, Y.; Zhou, M.; Shi, C.; French, N.P.; Shi, X. Identification and characterization of two novel superantigens among Staphylococcus aureus complex. Int. J. Med. Microbiol. 2018, 308, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.X.; McCormick, J.K. Staphylococcal superantigens in colonization and disease. Front. Cell. Infect. Microbiol. 2012, 2, 52. [Google Scholar] [CrossRef] [Green Version]
- Holtfreter, S.; Grumann, D.; Schmudde, M.; Nguyen, H.T.; Eichler, P.; Strommenger, B.; Kopron, K.; Kolata, J.; Giedrys-Kalemba, S.; Steinmetz, I.; et al. Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J. Clin. Microbiol. 2007, 45, 2669–2680. [Google Scholar] [CrossRef] [Green Version]
- Roetzer, A.; Haller, G.; Beyerly, J.; Geier, C.B.; Wolf, H.M.; Gruener, C.S.; Model, N.; Eibl, M.M. Genotypic and phenotypic analysis of clinical isolates of Staphylococcus aureus revealed production patterns and hemolytic potentials unlinked to gene profiles and source. BMC Microbiol. 2016, 16, 13. [Google Scholar] [CrossRef] [Green Version]
- Vu, B.G.; Stach, C.S.; Salgado-Pabón, W.; Diekema, D.J.; Gardner, S.E.; Schlievert, P.M. Superantigens of Staphylococcus aureus from patients with diabetic foot ulcers. J. Infect. Dis. 2014, 210, 1920–1927. [Google Scholar] [CrossRef] [Green Version]
- Fischer, A.J.; Kilgore, S.H.; Singh, S.B.; Allen, P.D.; Hansen, A.R.; Limoli, D.H.; Schlievert, P.M. High Prevalence of Staphylococcus aureus Enterotoxin Gene Cluster Superantigens in Cystic Fibrosis Clinical Isolates. Genes (Basel) 2019, 10, 1036. [Google Scholar] [CrossRef] [Green Version]
- Aung, M.S.; San, T.; Aye, M.M.; Mya, S.; Maw, W.W.; Zan, K.N.; Htut, W.H.W.; Kawaguchiya, M.; Urushibara, N.; Kobayashi, N. Prevalence and Genetic Characteristics of Staphylococcus aureus and Staphylococcus argenteus Isolates Harboring Panton-Valentine Leukocidin, Enterotoxins, and TSST-1 Genes from Food Handlers in Myanmar. Toxins (Basel) 2017, 9, 241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fooladvand, S.; Sarmadian, H.; Habibi, D.; van Belkum, A.; Ghaznavi-Rad, E. High prevalence of methicillin resistant and enterotoxin gene-positive Staphylococcus aureus among nasally colonized food handlers in central Iran. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Chao, G.; Bao, G.; Cao, Y.; Yan, W.; Wang, Y.; Zhang, X.; Zhou, L.; Wu, Y. Prevalence and diversity of enterotoxin genes with genetic background of Staphylococcus aureus isolates from different origins in China. Int. J. Food Microbiol. 2015, 211, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Mello, P.L.; Moraes Riboli, D.F.; Pinheiro, L.; de Almeida Martins, L.; Vasconcelos Paiva Brito, M.A.; Ribeiro de Souza da Cunha Mde, L. Detection of Enterotoxigenic Potential and Determination of Clonal Profile in Staphylococcus aureus and Coagulase-Negative Staphylococci Isolated from Bovine Subclinical Mastitis in Different Brazilian States. Toxins (Basel) 2016, 8, 104. [Google Scholar] [CrossRef] [Green Version]
- Shen, M.; Li, Y.; Zhang, L.; Dai, S.; Wang, J.; Li, Y.; Zhang, L.; Huang, J. Staphylococcus enterotoxin profile of China isolates and the superantigenicity of some novel enterotoxins. Arch. Microbiol. 2017, 199, 723–736. [Google Scholar] [CrossRef]
- Naimi, T.S.; LeDell, K.H.; Como-Sabetti, K.; Borchardt, S.M.; Boxrud, D.J.; Etienne, J.; Johnson, S.K.; Vandenesch, F.; Fridkin, S.; O’Boyle, C.; et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 2003, 290, 2976–2984. [Google Scholar] [CrossRef] [Green Version]
- Hu, D.L.; Omoe, K.; Inoue, F.; Kasai, T.; Yasujima, M.; Shinagawa, K.; Nakane, A. Comparative prevalence of superantigenic toxin genes in meticillin-resistant and meticillin-susceptible Staphylococcus aureus isolates. J. Med. Microbiol. 2008, 57, 1106–1112. [Google Scholar] [CrossRef] [Green Version]
- Shukla, S.K.; Karow, M.E.; Brady, J.M.; Stemper, M.E.; Kislow, J.; Moore, N.; Wroblewski, K.; Chyou, P.H.; Warshauer, D.M.; Reed, K.D.; et al. Virulence genes and genotypic associations in nasal carriage, community-associated methicillin-susceptible and methicillin-resistant USA400 Staphylococcus aureus isolates. J. Clin. Microbiol. 2010, 48, 3582–3592. [Google Scholar] [CrossRef] [Green Version]
- Ono, H.K.; Sato’o, Y.; Narita, K.; Naito, I.; Hirose, S.; Hisatsune, J.; Asano, K.; Hu, D.L.; Omoe, K.; Sugai, M.; et al. Identification and Characterization of a Novel Staphylococcal Emetic Toxin. Appl. Environ. Microbiol. 2015, 81, 7034–7040. [Google Scholar] [CrossRef] [Green Version]
- Aziz, F.; Hisatsune, J.; Yu, L.; Kajimura, J.; Sato’o, Y.; Ono, H.K.; Masuda, K.; Yamaoka, M.; Salasia, S.I.O.; Nakane, A.; et al. Staphylococcus aureus Isolated from Skin from Atopic-Dermatitis Patients Produces Staphylococcal Enterotoxin Y, Which Predominantly Induces T-Cell Receptor Vα-Specific Expansion of T Cells. Infect. Immun. 2020, 88, e00360–e00419. [Google Scholar] [CrossRef] [Green Version]
- Wilson, G.J.; Seo, K.S.; Cartwright, R.A.; Connelley, T.; Chuang-Smith, O.N.; Merriman, J.A.; Guinane, C.M.; Park, J.Y.; Bohach, G.A.; Schlievert, P.M.; et al. A novel core genome-encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathog. 2011, 7, e1002271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langley, R.J.; Ting, Y.T.; Clow, F.; Young, P.G.; Radcliff, F.J.; Choi, J.M.; Sequeira, R.P.; Holtfreter, S.; Baker, H.; Fraser, J.D. Staphylococcal enterotoxin-like X (SElX) is a unique superantigen with functional features of two major families of staphylococcal virulence factors. PLoS Pathog. 2017, 13, e1006549. [Google Scholar] [CrossRef] [PubMed]
- Tuffs, S.W.; James, D.B.A.; Bestebroer, J.; Richards, A.C.; Goncheva, M.I.; O’Shea, M.; Wee, B.A.; Seo, K.S.; Schlievert, P.M.; Lengeling, A.; et al. The Staphylococcus aureus superantigen SElX is a bifunctional toxin that inhibits neutrophil function. PLoS Pathog. 2017, 13, e1006461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okumura, K.; Shimomura, Y.; Murayama, S.Y.; Yagi, J.; Ubukata, K.; Kirikae, T.; Miyoshi-Akiyama, T. Evolutionary paths of streptococcal and staphylococcal superantigens. BMC Genom. 2012, 13, 404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lina, G.; Bohach, G.A.; Nair, S.P.; Hiramatsu, K.; Jouvin-Marche, E.; Mariuzza, R. International Nomenclature Committee for Staphylococcal Superantigens. Standard nomenclature for the superantigens expressed by Staphylococcus. J. Infect. Dis. 2004, 189, 2334–2336. [Google Scholar] [CrossRef] [Green Version]
- Thomas, D.Y.; Jarraud, S.; Lemercier, B.; Cozon, G.; Echasserieau, K.; Etienne, J.; Gougeon, M.L.; Lina, G.; Vandenesch, F. Staphylococcal enterotoxin-like toxins U2 and V, two new staphylococcal superantigens arising from recombination within the enterotoxin gene cluster. Infect. Immun. 2006, 74, 4724–4734. [Google Scholar] [CrossRef] [Green Version]
- Collery, M.M.; Smyth, C.J. Rapid differentiation of Staphylococcus aureus isolates harbouring egc loci with pseudogenes ψent1 and ψent2 and the selu or seluv gene using PCR-RFLP. J. Med. Microbiol. 2007, 56, 208–216. [Google Scholar] [CrossRef] [Green Version]
- Argudín, M.Á.; Mendoza, M.C.; Rodicio, M.R. Food poisoning and Staphylococcus aureus enterotoxins. Toxins (Basel) 2010, 2, 1751–1773. [Google Scholar] [CrossRef]
- Fisher, E.L.; Otto, M.; Cheung, G.Y.C. Basis of Virulence in Enterotoxin-Mediated Staphylococcal Food Poisoning. Front. Microbiol. 2018, 9, 436. [Google Scholar] [CrossRef]
- Benkerroum, N. Staphylococcal enterotoxins and enterotoxin-like toxins with special reference to dairy products: An overview. Crit. Rev. Food Sci. Nutr. 2018, 58, 1943–1970. [Google Scholar] [CrossRef]
- Tuffs, S.W.; Haeryfar, S.M.M.; McCormick, J.K. Manipulation of Innate and Adaptive Immunity by Staphylococcal Superantigens. Pathogens 2018, 7, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chieffi, D.; Fanelli, F.; Chob, G.S.; Schubert, J.; Blaiotta, G.; Franz, C.M.A.P.; Bania, J.; Fusco, V. Novel insights into the enterotoxigenic potential and genomic background of Staphylococcus aureus isolated from raw milk. Food Microbiol. 2020, 90, 103482. [Google Scholar] [CrossRef] [PubMed]
- Aung, M.S.; Kawaguchiya, M.; Urushibara, N.; Sumi, A.; Ito, M.; Kudo, K.; Morimoto, S.; Hosoya, S.; Kobayashi, N. Molecular Characterization of Methicillin-Resistant Staphylococcus aureus from Outpatients in Northern Japan: Increasing Tendency of ST5/ST764 MRSA-IIa with Arginine Catabolic Mobile Element. Microb. Drug. Resist. 2017, 23, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Aung, M.S.; Urushibara, N.; Kawaguchiya, M.; Sumi, A.; Takahashi, S.; Ike, M.; Ito, M.; Habadera, S.; Kobayashi, N. Molecular Epidemiological Characterization of Staphylococcus argenteus Clinical Isolates in Japan: Identification of Three Clones (ST1223, ST2198, and ST2550) and a Novel Staphylocoagulase Genotype XV. Microorganisms 2019, 7, 389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, Q.; Shang, W.; Hu, X.; Rao, X. Staphylococcus aureus ST121: A globally disseminated hypervirulent clone. J. Med. Microbiol. 2015, 64, 1462–1473. [Google Scholar] [CrossRef]
- Kawaguchiya, M.; Urushibara, N.; Kuwahara, O.; Ito, M.; Mise, K.; Kobayashi, N. Molecular characteristics of community-acquired methicillin-resistant Staphylococcus aureus in Hokkaido, northern main island of Japan: Identification of sequence types 6 and 59 Panton-Valentine leucocidin-positive community-acquired methicillin-resistant Staphylococcus aureus. Microb. Drug Resist. 2011, 17, 241–250. [Google Scholar]
- Kawaguchiya, M.; Urushibara, N.; Yamamoto, D.; Yamashita, T.; Shinagawa, M.; Watanabe, N.; Kobayashi, N. Characterization of PVL/ACME-positive methicillin-resistant Staphylococcus aureus (genotypes ST8-MRSA-IV and ST5-MRSA-II) isolated from a university hospital in Japan. Microb. Drug Resist. 2013, 19, 48–56. [Google Scholar] [CrossRef]
- Aung, M.S.; Urushibara, N.; Kawaguchiya, M.; Sumi, A.; Shinagawa, M.; Takahashi, S.; Kobayashi, N. Clonal Diversity and Genetic Characteristics of Methicillin-Resistant Staphylococcus aureus Isolates from a Tertiary Care Hospital in Japan. Microb. Drug Resist. 2019, 25, 1164–1175. [Google Scholar] [CrossRef]
- Hirose, M.; Kobayashi, N.; Ghosh, S.; Paul, S.K.; Shen, T.; Urushibara, N.; Kawaguchiya, M.; Shinagawa, M.; Watanabe, N. Identification of staphylocoagulase genotypes I-X and discrimination of type IV and V subtypes by multiplex PCR assay for clinical isolates of Staphylococcus aureus. Jpn. J. Infect. Dis. 2010, 63, 257–263. [Google Scholar]
- Aung, M.S.; Urushibara, N.; Kawaguchiya, M.; Aung, T.S.; Mya, S.; San, T.; New, K.M.; Kobayashi, N. Virulence factors and genetic characteristics of methicillin-resistant and -susceptible Staphylococcus aureus isolates in Myanmar. Microb. Drug Resist. 2011, 17, 525–535. [Google Scholar] [CrossRef] [Green Version]


| Genotype | Total No. of Isolates | No. of Isolates With SE(-Like) Gene *1 (%) | ||||
|---|---|---|---|---|---|---|
| coa Genotype | spa Type (n = 149) *2 | sey | selw | selx | selz | |
| Ia | t375 (3) | 17 | 8 (47.1) | 8 (47.1) | 13 (76.5) | 12 (70.6) |
| IIa | t002 (56), t548 (2), t2487 (2), t001 (1), t045 (2) | 455 | 157 (34.5) | 450 (98.9) | 397 (87.3) | 5 (1.1) |
| IIIa | t008 (9), t4133 (2), t1767 (24), t5071 (1), t1627 (2), t1581(2) | 74 | 22 (29.7) | 64 (86.5) | 60 (81.1) | 0 |
| IVa | t019 (1) | 3 | 0 | 1 (33.3) | 0 | 0 |
| Va | t5110 (6), t10641 (10) | 16 | 13 (81.3) | 8 (50) | 16 (100) | 14 (87.5) |
| Vb | NT (1) | 1 | 0 | 1 (100) | 1 (100) | 0 |
| VIa | ND | 4 | 0 | 0 | 0 | 0 |
| VIIa | t1784 (23) | 51 | 20 (39.2) | 46 (90.2) | 46 (90.2) | 4 (7.8) |
| VIIb | t370 (2) | 3 | 1 (33.3) | 2 (66.7) | 1 (33.3) | 0 |
| Total | 624 | 221 (35.4) | 580 (92.9) | 534 (85.6) | 35 (5.6) | |
| SCCmec type | ||||||
| SCCmec I | 2 | 0 | 1 (50) | 2 (100) | 0 | |
| SCCmec II | 452 | 154 (34.1) | 431 (95.4) | 393 (86.9) | 5 (1.1) | |
| SCCmec IV | 125 | 41 (32.8) | 115 (92) | 106 (84.8) | 4 (3.2) | |
| SCCmec V | 34 | 26 (76.5) | 30 (88.2) | 30 (88.2) | 26 (76.5) | |
| SCCmec NT | 11 | 0 | 3 (27.3) | 3 (27.3) | 0 | |
| Specimen | ||||||
| sputum | 136 | 45 (33.1) | 130 (95.6) | 121 (89) | 9 (6.6) | |
| urine | 129 | 40 (31) | 118 (91.5) | 102 (79.1) | 3 (2.3) | |
| ear discharge | 76 | 35 (46.1) | 72 (94.7) | 70 (92.1) | 6 (7.9) | |
| nasal discharge | 75 | 28 (37.3) | 73 (97.3) | 67 (89.3) | 7 (9.3) | |
| pus | 57 | 24 (42.1) | 54 (94.7) | 52 (91.2) | 3 (5.3) | |
| wound swab | 29 | 13 (44.8) | 27 (93.1) | 27 (93.1) | 3 (10.3) | |
| eye swab | 29 | 12 (41.4) | 28 (96.6) | 26 (89.7) | 2 (6.9) | |
| stool | 33 | 9 (27.3) | 26 (78.8) | 24 (72.7) | 0 | |
| skin | 26 | 10 (38.5) | 24 (92.3) | 23 (88.5) | 2 (7.7) | |
| Others *3 | 34 | 5 (14.7) | 28 (82.4) | 22 (64.7) | 0 | |
| PVL/ACME Genes | ST (CC) | Total No. of Isolates (n = 100) | SE(-Like) Genes Identified *2 |
|---|---|---|---|
| PVL+/ACME+ | ST8 (CC8) | 9 *1 | sek, seq, selw, selx |
| PVL+/ACME- | ST30 (CC30) | 1 *1 | sem, sen, seo, seu, selw, selx |
| ST59 (CC59) | 1 *1 | seb, sek, seq, selw, selx | |
| PVL-/ACME+ | ST5/ST764 (CC5) | 15 *1 | seb (67%), sec (20%), seg, sei, sem, sen, seo, seu, sep (33%), selw, selx, sey |
| PVL-/ACME- | ST8 (CC8) (CA-MRSA/J *3) | 5 *1 | sec, sel, sep, selw, selx |
| ST8 (CC8) | 14 | selj (29%), ser (29%), selw (93%), selx (93%), sey (43%) | |
| ST5/ST764 (CC5) | 20 | seb (60%), sec (10%), seg, sei, sem, sen, seo, seu, selw, selx, sey (30%), selz (25%) | |
| ST5425 (CC5) | 1 | seg, sei, sem, sen, seo, selw, selx | |
| ST45 (CC45) | 2 | seg, sei, sem, sen, seo, selw, selx, sey | |
| ST1 (CC1) | 13 | sea, sek, seq, selx (92%), selw (92%), sey (31%), selz (15%) | |
| ST89 (CC89) | 8 | sem, seo, seu, selw, selx, sey, selz | |
| ST121 (CC121) | 10 | seg, sei, sem, sen, seo, seu, selw (50%), selx, sey (80%), selz (85%) | |
| ST12 (CC12) | 1 | sep, selw, selx, selz |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aung, M.S.; Urushibara, N.; Kawaguchiya, M.; Ito, M.; Habadera, S.; Kobayashi, N. Prevalence and Genetic Diversity of Staphylococcal Enterotoxin (-Like) Genes sey, selw, selx, selz, sel26 and sel27 in Community-Acquired Methicillin-Resistant Staphylococcus aureus. Toxins 2020, 12, 347. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12050347
Aung MS, Urushibara N, Kawaguchiya M, Ito M, Habadera S, Kobayashi N. Prevalence and Genetic Diversity of Staphylococcal Enterotoxin (-Like) Genes sey, selw, selx, selz, sel26 and sel27 in Community-Acquired Methicillin-Resistant Staphylococcus aureus. Toxins. 2020; 12(5):347. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12050347
Chicago/Turabian StyleAung, Meiji Soe, Noriko Urushibara, Mitsuyo Kawaguchiya, Masahiko Ito, Satoshi Habadera, and Nobumichi Kobayashi. 2020. "Prevalence and Genetic Diversity of Staphylococcal Enterotoxin (-Like) Genes sey, selw, selx, selz, sel26 and sel27 in Community-Acquired Methicillin-Resistant Staphylococcus aureus" Toxins 12, no. 5: 347. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12050347





