Production of Paralytic Shellfish Toxins (PSTs) in Toxic Alexandrium catenella is Intertwined with Photosynthesis and Energy Production
Abstract
:1. Introduction
2. Results
2.1. DNA-Based Identification
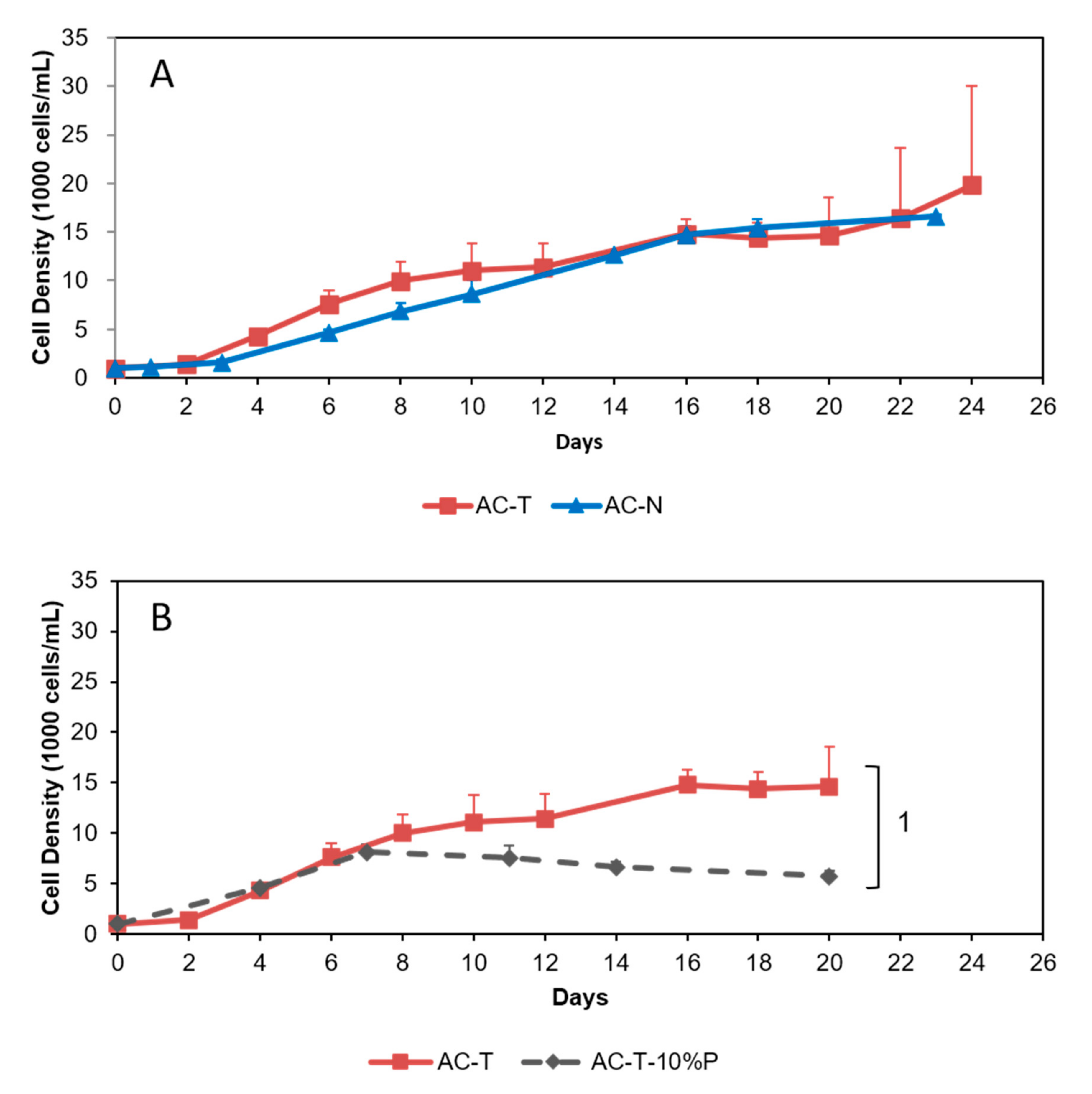
2.2. Growth Curves
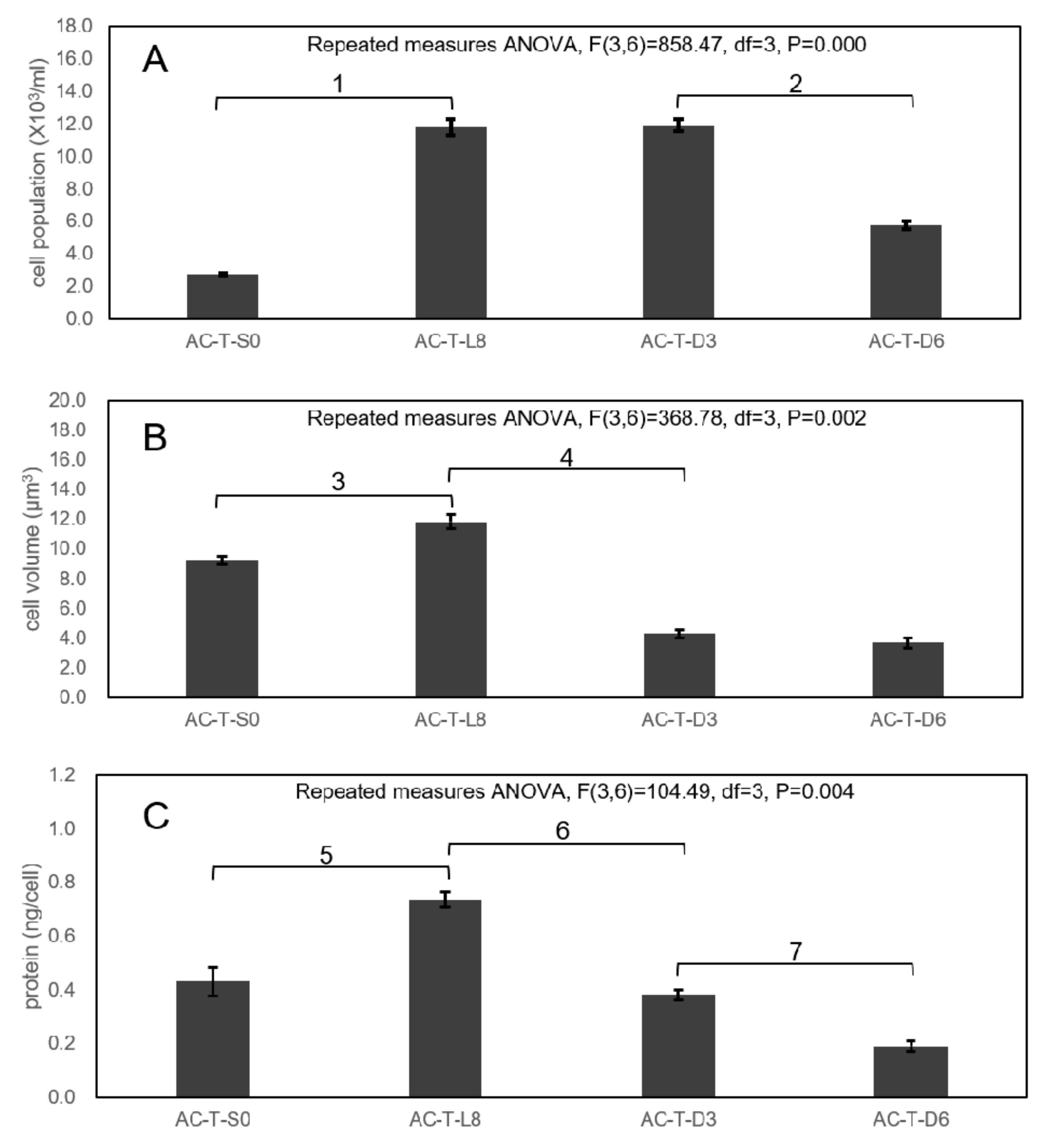
2.3. Cell Sizes of AC-N, AC-T, and AC-T with Phosphate Limitation
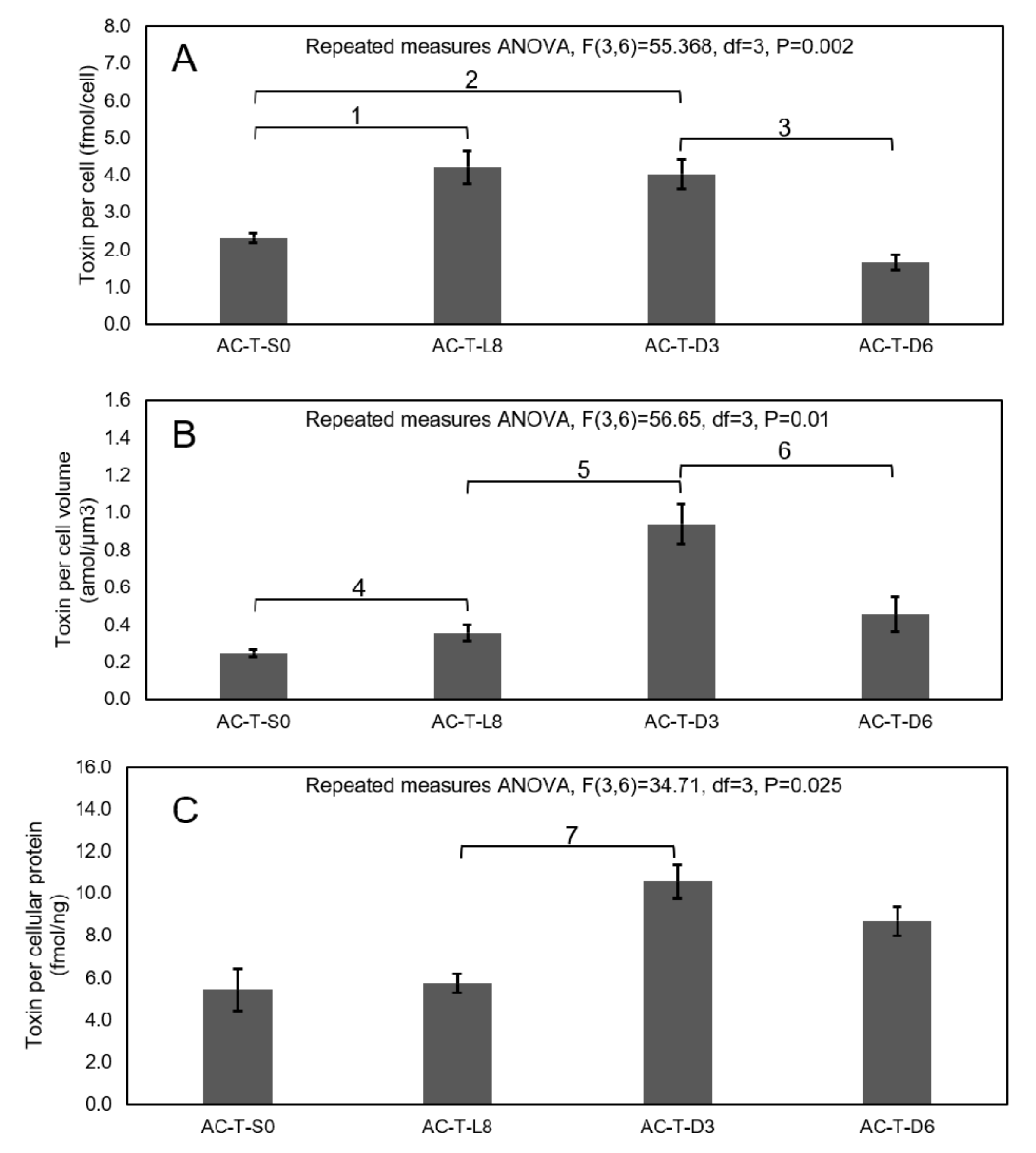
2.4. Comparison of Toxicity of AC-N, AC-T, and AC-T with Phosphate Limitation
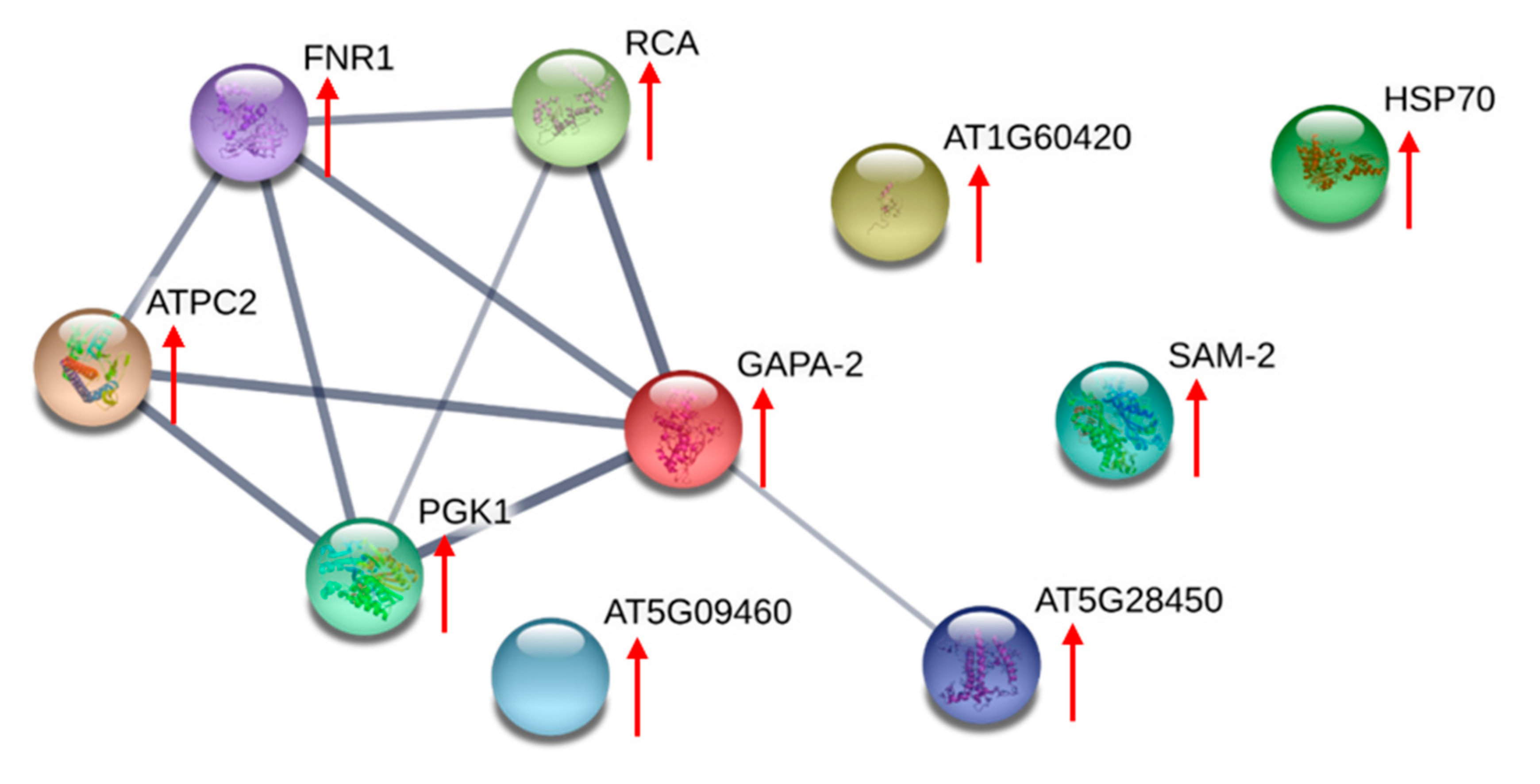
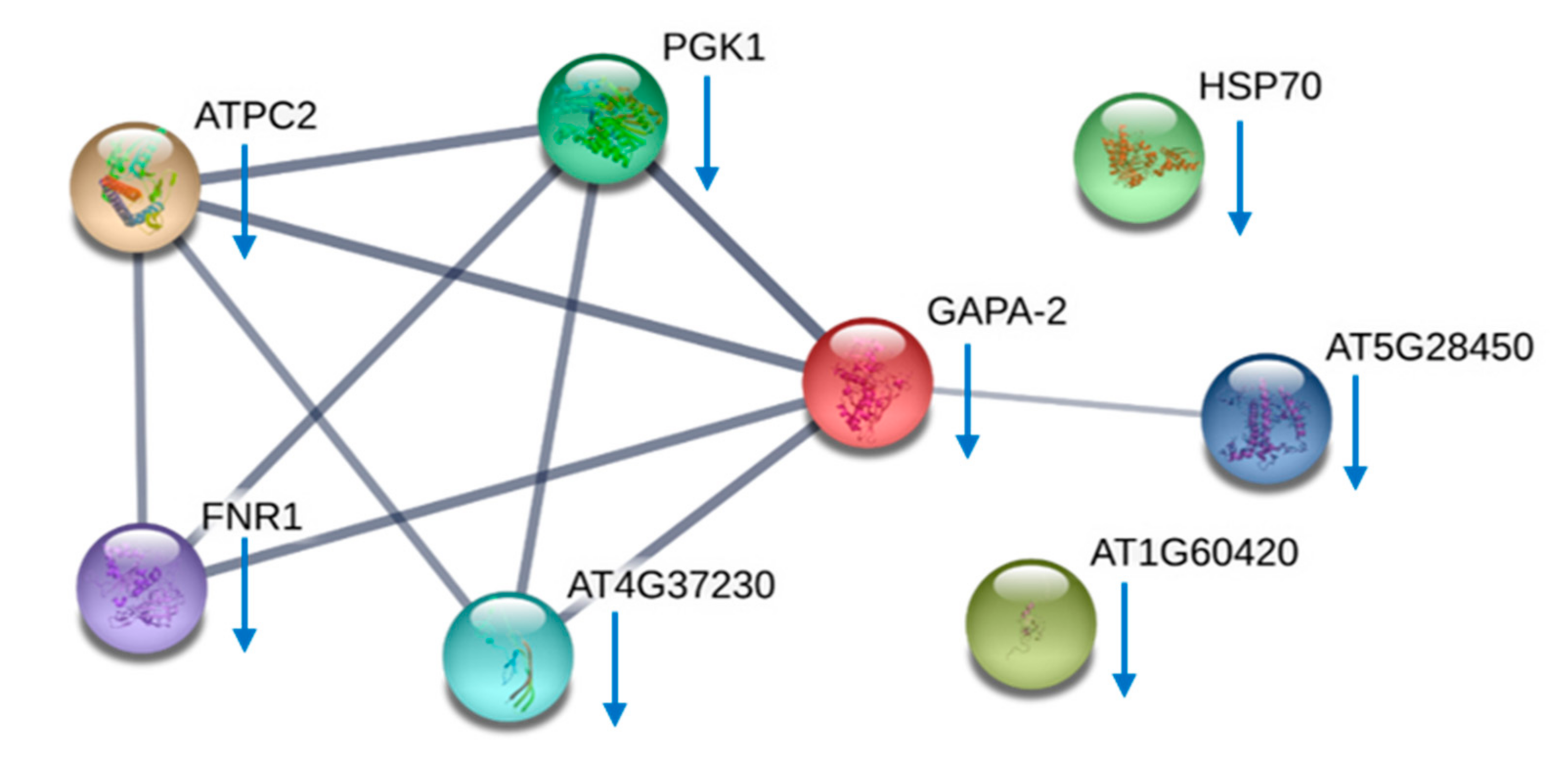
2.5. Proteomes Comparison with 2-Dimensional Gel Electrophoresis (2-DE) Profiles and Protein Identification
2.6. Photoperiod Experiment to Investigate Effects on Toxicity of AC-T
3. Discussion
4. Materials and Methods
4.1. Chemicals and Materials
4.2. Dinoflagellates Cultures and Cultivating Conditions
4.3. DNA-Based Identification
4.4. Cell Counts and Growth Curves
4.5. Cell Size Measurement
4.6. Cellular Protein Analysis
4.7. Toxin Analysis
4.8. Protein Extraction and 2-Dimensional Gel Electrophoresis (2-DE)
4.9. Gel Staining and Image Analysis
4.10. In-Gel Digestion
4.11. MALDI-TOF-MS Analysis and N-terminal Sulfonation
4.12. LC-ESI-iontrap Analysis
4.13. STRING Analyses
4.14. Photoperiod Experiments
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Garcıa, C.; del Carmen Bravo, M.; Lagos, M.; Lagos, N. Paralytic shellfish poisoning: Post-mortem analysis of tissue and body fluid samples from human victims in the Patagonia fjords. Toxicon 2004, 43, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Sobel, J.; Painter, J. Illnesses caused by marine toxins. Clin. Infect. Dis. 2005, 41, 1290–1296. [Google Scholar] [CrossRef] [PubMed]
- Deeds, J.R.; Landsberg, J.H.; Etheridge, S.M.; Pitcher, G.C.; Longan, S.W. Non-traditional vectors for paralytic shellfish poisoning. Mar. Drugs 2008, 6, 308–348. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.-T.; Ogata, T. Salinity effect on growth and toxin production of four tropical Alexandrium species (Dinophyceae). Toxicon 2005, 45, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Ichimi, K.; Suzuki, T.; Ito, A. Variety of PSP toxin profiles in various culture strains of Alexandrium tamarense and change of toxin profile in natural A. tamarense population. J. Exp. Mar. Biol. Ecol. 2002, 273, 51–60. [Google Scholar] [CrossRef]
- Caruana, A.M.; Amzil, Z. Microalgae and Toxins. In Microalgae in Health and Disease Prevention; Elsevier: Amsterdam, The Netherlands, 2018; pp. 263–305. [Google Scholar]
- Boyer, G.; Sullivan, J.; Andersen, R.; Harrison, P.; Taylor, F. Effects of nutrient limitation on toxin production and composition in the marine dinoflagellate Protogonyaulax tamarensis. Mar. Biol. 1987, 96, 123–128. [Google Scholar] [CrossRef]
- Anderson, D.; Kulis, D.; Sullivan, J.; Hall, S. Toxin composition variations in one isolate of the dinoflagellate Alexandrium fundyense. Toxicon 1990, 28, 885–893. [Google Scholar] [CrossRef]
- Lippemeier, S.; Frampton, D.M.; Blackburn, S.I.; Geier, S.C.; Negri, A.P. Influence of phosphorus limitation on toxicity and photosynthesis of Alexandrium minutum (Dinophyceae) moniteres by in-line detection of variable chlorophyll flourescence 1. J. Phycol. 2003, 39, 320–331. [Google Scholar] [CrossRef]
- Leong, S.C.Y.; Murata, A.; Nagashima, Y.; Taguchi, S. Variability in toxicity of the dinoflagellate Alexandrium tamarense in response to different nitrogen sources and concentrations. Toxicon 2004, 43, 407–415. [Google Scholar] [CrossRef]
- Lim, P.-T.; Leaw, C.-P.; Kobiyama, A.; Ogata, T. Growth and toxin production of tropical Alexandrium minutum Halim (Dinophyceae) under various nitrogen to phosphorus ratios. J. Appl. Phycol. 2010, 22, 203–210. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Ho, A.Y.; He, L.; Yin, K.; Hung, C.; Choi, N.; Lam, P.K.; Wu, R.S.; Anderson, D.M.; Harrison, P.J. Effects of inorganic and organic nitrogen and phosphorus on the growth and toxicity of two Alexandrium species from Hong Kong. Harmful Algae 2012, 16, 89–97. [Google Scholar] [CrossRef]
- Van de Waal, D.B.; Smith, V.H.; Declerck, S.A.; Stam, E.C.; Elser, J.J. Stoichiometric regulation of phytoplankton toxins. Ecol. Lett. 2014, 17, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Kellmann, R.; Michali, T.K.; Neilan, B.A. Identification of a saxitoxin biosynthesis gene with a history of frequent horizontal gene transfers. J. Mol. Evol. 2008, 67, 526–538. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, A.; Loram, J.E.; Hackett, J.D.; Anderson, D.M.; Plumley, F.G.; Bhattacharya, D. Origin of saxitoxin biosynthetic genes in cyanobacteria. PLoS ONE 2009, 4, 5758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stüken, A.; Orr, R.J.; Kellmann, R.; Murray, S.A.; Neilan, B.A.; Jakobsen, K.S. Discovery of nuclear-encoded genes for the neurotoxin saxitoxin in dinoflagellates. PLoS ONE 2011, 6, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taroncher-Oldenburg, G.; Anderson, D.M. Identification and characterization of three differentially expressed genes, encoding S-adenosylhomocysteine hydrolase, methionine aminopeptidase, and a histone-like protein, in the toxic dinoflagellate Alexandrium fundyense. Appl. Environ. Microbiol. 2000, 66, 2105–2112. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.; Ogawa, M.; Hirota, M.; Oshima, Y. Effects of mitomycin C and colchicine on toxin production and cell cycle regulation in the dinoflagellate Alexandrium tamarense. Harmful Algae 2011, 10, 235–244. [Google Scholar] [CrossRef]
- Zhang, S.-F.; Zhang, Y.; Lin, L.; Wang, D.-Z. iTRAQ-based quantitative proteomic analysis of a toxigenic dinoflagellate Alexandrium catenella and its non-toxigenic mutant exposed to a cell cycle inhibitor colchicine. Front. Microbiol. 2018, 9, 650. [Google Scholar] [CrossRef]
- Le Gac, M.; Metegnier, G.; Chomérat, N.; Malestroit, P.; Quéré, J.; Bouchez, O.; Siano, R.; Destombe, C.; Guillou, L.; Chapelle, A. Evolutionary processes and cellular functions underlying divergence in Alexandrium minutum. Mol. Ecol. 2016, 25, 5129–5143. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.; Ogawa, M.; Yotsu-Yamashita, M.; Oshima, Y. Effect of 5-fluoro-2′-deoxyuridine on toxin production and cell cycle regulation in marine dinoflagellate, Alexandrium tamarense. Harmful Algae 2014, 32, 64–72. [Google Scholar] [CrossRef]
- Wang, D.-Z.; Li, C.; Zhang, Y.; Wang, Y.-Y.; He, Z.-P.; Lin, L.; Hong, H.-S. Quantitative proteomic analysis of differentially expressed proteins in the toxicity-lost mutant of Alexandrium catenella (Dinophyceae) in the exponential phase. J. Proteom. 2012, 75, 5564–5577. [Google Scholar] [CrossRef]
- Zhang, S.F.; Zhang, Y.; Xie, Z.X.; Zhang, H.; Lin, L.; Wang, D.Z. iTRAQ-based quantitative proteomic analysis of a toxigenic dinoflagellate Alexandrium catenella and its non-toxic mutant. Proteomics 2015, 15, 4041–4050. [Google Scholar] [CrossRef]
- Martins, C.A.; Kulis, D.; Franca, S.; Anderson, D.M. The loss of PSP toxin production in a formerly toxic Alexandrium lusitanicum clone. Toxicon 2004, 43, 195–205. [Google Scholar] [CrossRef]
- John, U.; Litaker, R.W.; Montresor, M.; Murray, S.; Brosnahan, M.L.; Anderson, D.M. Formal revision of the Alexandrium tamarense species complex (Dinophyceae) taxonomy: The introduction of five species with emphasis on molecular-based (rDNA) classification. Protist 2014, 165, 779–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, D.; Kulis, D.; Sullivan, J.; Hall, S.; Lee, C. Dynamics and physiology of saxitoxin production by the dinoflagellates Alexandrium spp. Mar. Biol. 1990, 104, 511–524. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2017, 45, D37–D42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hillebrand, H.; Dürselen, C.D.; Kirschtel, D.; Pollingher, U.; Zohary, T. Biovolume calculation for pelagic and benthic microalgae. J. Phycol. 1999, 35, 403–424. [Google Scholar] [CrossRef]
- Wang, D.-Z.; Gao, Y.; Lin, L.; Hong, H.-S. Comparative proteomic analysis reveals proteins putatively involved in toxin biosynthesis in the marine dinoflagellate Alexandrium catenella. Mar. Drugs 2013, 11, 213–232. [Google Scholar] [CrossRef] [Green Version]
- Lee, F.W.-F.; Morse, D.; Lo, S.C.-L. Identification of two plastid proteins in the dinoflagellate Alexandrium affine that are substantially down-regulated by nitrogen-depletion. J. Proteome Res. 2009, 8, 5080–5092. [Google Scholar] [CrossRef]
- Zhuang, Y.; Zhang, H.; Hannick, L.; Lin, S. Metatranscriptome profiling reveals versatile N-nutrient utilization, CO2 limitation, oxidative stress, and active toxin production in an Alexandrium fundyense bloom. Harmful Algae 2015, 42, 60–70. [Google Scholar] [CrossRef]
- Shimizu, Y. Microalgal metabolites. Chem. Rev. 1993, 93, 1685–1698. [Google Scholar] [CrossRef]
- Neilan, B.A. Current knowledge of paralytic shellfish toxin biosynthesis, molecular detection and evolution. Toxins Biol. Act. Compd. Microalgae 2014, 1, 251. [Google Scholar]
- Zhang, S.-F.; Zhang, Y.; Lin, L.; Wang, D.-Z. iTRAQ-based quantitative proteomic analysis of a toxigenic dinoflagellate Alexandrium catenella at different stages of toxin biosynthesis during the cell cycle. Mar. Drugs 2018, 16, 491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maas, E.W.; Brooks, H.J.L. Is photosynthesis a requirement for paralytic shellfish toxin production in the dinoflagellate Alexandrium minutum algal–bacterial consortium? J. Appl. Phycol. 2010, 22, 293–296. [Google Scholar] [CrossRef]
- Zhang, S.-F.; Chen, Y.; Xie, Z.-X.; Zhang, H.; Lin, L.; Wang, D.-Z. Unraveling the molecular mechanism of the response to changing ambient phosphorus in the dinoflagellate Alexandrium catenella with quantitative proteomics. J. Proteom. 2019, 196, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.D.; Selvin, R.C.; Claus, W.; Guillard, R.R. Media for the culture of oceanic ultraphytoplankton1, 2. J. Phycol. 1987, 23, 633–638. [Google Scholar] [CrossRef]
- Adachi, M.; Sake, Y.; Ishida, Y. Analysis of Alexandrium (Dinophyceae) species using sequences of the 5.8 S ribosomal DNA and internal transcribed spacer regions 1. J. Phycol. 1996, 32, 424–432. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 1, 2–3. [Google Scholar] [CrossRef]
- Altschul, S.; Gish, W.; Miller, W.; Myers, E.; Lipman, D. Basic local alignment search tool. J. Mol. Biol. 1990, 215. [Google Scholar] [CrossRef]
- Coordinators, N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, J.F.; Wong, B.; Ménard, C. Determination of decarbamoyl saxitoxin and its analogues in shellfish by prechromatographic oxidation and liquid chromatography with fluorescence detection. J. AOAC Int. 1996, 79, 1111–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, F.W.-F.; Lo, S.C.-L. The use of Trizol reagent (phenol/guanidine isothiocyanate) for producing high quality two-dimensional gel electrophoretograms (2-DE) of dinoflagellates. J. Microbiol. Methods 2008, 73, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Kalb, S.R.; Cotter, R.J. Improved procedures for N-terminal sulfonation of peptides for matrix-assisted laser desorption/ionization post-source decay peptide sequencing. Rapid Commun. Mass Spectrom. 2004, 18, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Howell, D.C. Statistical Methods for Psychology; Cengage Learning: Belmont, CA, USA, 2009. [Google Scholar]
- Field, A. Discovering Statistics Using IBM SPSS Statistics; Sage: Thousand Oaks, CA, USA, 2013. [Google Scholar]





| Significantly Matched Proteins | Differential Expression Ratio (AC-T: AC-N) | NCBI Accession no. | Spot Number | pI Value (pI) | Molecular Weight (kDa) | Mascot Score | Identified by |
|---|---|---|---|---|---|---|---|
| (More abundant in AC-T) | |||||||
| ribulose 1,5-bisphosphate carboxylase/oxygenase II | 5.2:1 | AAO13045.1 AAO13070.1 | 27 | 5.4 | 60 | Blastp E-value: 1.00E-04 (de novo sequencing) | MALDI-TOF |
| 5.2:1 | 28 | 5.4 | 60 | ||||
| 4.7:1 | 31 | 5.5 | 60 | ||||
| 4.1:1 | 32 | 5.6 | 60 | ||||
| 3.2:1 | 33 | 5.7 | 60 | ||||
| methionine adenosyltransferase | 5.9:1 | NP_001035469.1 | 38 | 6.6 | 60 | 141 | MALDI-TOF |
| chloroplast ATP synthase gamma submit | 2.5:1 | AAW79296.1 | 54 | 5.4 | 55 | 197 | ESI-iontrap |
| nitrogen associated protein 50 | 2.2:1 | ACV60537.1 | 63 | 5.4 | 50 | 232 | ESI-iontrap |
| 2.2:1 | 65 | 5.5 | 50 | ||||
| nucleoredoxin | Only detected in AC-T | KOO32230.1 | 73 | 5.0 | 48 | 167 | ESI-iontrap |
| phosphoglycerate kinase | 5.5:1 | OLP97413.1 | 96 | 5.3 | 43 | 332 | ESI-iontrap |
| 4.5:1 | 97 | 5.4 | 43 | ||||
| 2.9:1 | 98 | 5.5 | 43 | ||||
| glyceraldehyde-3 phosphate dehydrogenase | 7.9:1 | ABI14256.1 | 104 | 6.5 | 42 | 110 | MALDI-TOF |
| chloroplast ferredoxin-NADP(+) reductase | 4.7:1 | AAW79314.1 | 117 | 6.4 | 42 | 461 | ESI-iontrap |
| 7.0:1 | 118 | 6.6 | 42 | ||||
| 8.2:1 | 123 | 6.7 | 42 | ||||
| peridinin chlorophyll-a binding protein apoprotein precursor | 3.0:1 | AFH88375.1 | 175 | 6.5 | 30 | 1343 | ESI-iontrap |
| response regulator | 2.4:1 | WP_011605320.1 | 257 | 5.0 | 19 | 67 | MALDI-TOF |
| light harvesting protein | 2.0:1 | CBI83417.1 | 266 | 5.0 | 19 | 650 | ESI-iontrap |
| heat shock protein 70 | 6.1:1 | AAM02973.2 | 10 | 5.2 | 80 | 487 | ESI-iontrap |
| 7.7:1 | 15 | 5.3 | 78 | ||||
| 6.5:1 | 14 | 5.2 | 78 | ||||
| Significantly Matched Proteins | Differential Expression Ratio (AC-T:AC-T-10%P) | NCBI Accession No. | Spot Number | pI Value (pI) | Molecular Weight (kDa) | Mascot Score | Identified by |
|---|---|---|---|---|---|---|---|
| (More abundant in AC-T) | |||||||
| heat shock protein 70 | 1:3.7 | AAM02973.2 | 10 | 5.2 | 80 | 487 | ESI-iontrap |
| 1:7.2 | 15 | 5.3 | 78 | ||||
| 1:2.5 | 14 | 5.2 | 78 | ||||
| chloroplast ATP synthase | 1:3.7 | AAW79296.1 | 54 | 5.4 | 55 | 197 | ESI-iontrap |
| nitrogen associated protein 50 | 1:5.9 | ACV605371. | 63 | 5.4 | 50 | 232 | ESI-iontrap |
| 1:2.1 | 65 | 5.5 | 50 | ||||
| 1:2.0 | 66 | 5.5 | 50 | ||||
| nucleoredoxin | 1:3.7 | KOO32230.1 | 73 | 5.0 | 48 | 167 | ESI-iontrap |
| phosphoglycerate kinase | 1:7.3 | OLP97413.1 | 96 | 5.3 | 43 | 332 | ESI-iontrap |
| 1:7.7 | 97 | 5.4 | 43 | ||||
| 1:6.1 | 98 | 5.5 | 43 | ||||
| 10lyceraldehyde-3 phosphate dehydrogenase | 1:2.9 | ABI14256.1 | 104 | 6.5 | 43 | 110 | MALDI-TOF |
| chloroplast ferredoxin-NADP(+) reductase | 1:3.0 | AAW79314.1 | 117 | 6.4 | 42 | 461 | ESI-iontrap |
| 1:2.9 | 118 | 6.6 | 42 | ||||
| 1:2.4 | 123 | 6.7 | 42 | ||||
| peridinin chlorophyll-a binding protein | 1:4.2 | AFH88375.1 | 192 | 6.3 | 28 | 407 | ESI-iontrap |
| oxygen-evolving enhancer-1 protein | 1:3.5 | OLQ01973.1 | 196 | 6.8 | 29 | 334 | ESI-iontrap |
| light harvesting protein | 1:2.2 | AAM02973.2 | 266 | 5.0 | 19 | 650 | ESI-iontrap |
| Identified Proteins | General Functions | Differentially Expressed between AC-T and AC-N | Differentially Expressed between AC-T and AC-T-10%P |
|---|---|---|---|
| Ribulose 1,5-bisphosphate carboxylase/oxygenase II | Photosynthesis related | AC-T ↑ | Not differentially expressed |
| Chloroplast ATP synthase | AC-T-10%P ↓ | ||
| Chloroplast ferredoxin-NADP(+) reductase | |||
| Light-harvesting protein | |||
| Peridinin chlorophyll-a binding protein apoprotein precursor | |||
| Oxygen-evolving enhancer-1 protein | Not differentially expressed | ||
| Glyceraldehydes-3-phosphate dehydrogenase | Cellular energy-related | AC-T ↑ | AC-T-10%P ↓ |
| Phosphoglycerate kinase | |||
| Nitrogen associated protein 50 | Unknown | ||
| Nucleoredoxin | Signaling related | ||
| Heat shock protein 70 | Environmental stress-related |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tse, S.P.-k.; Lee, F.W.-f.; Mak, D.Y.-l.; Kong, H.-k.; Chan, K.K.-y.; Lo, P.-y.; Lo, S.C.-l. Production of Paralytic Shellfish Toxins (PSTs) in Toxic Alexandrium catenella is Intertwined with Photosynthesis and Energy Production. Toxins 2020, 12, 477. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12080477
Tse SP-k, Lee FW-f, Mak DY-l, Kong H-k, Chan KK-y, Lo P-y, Lo SC-l. Production of Paralytic Shellfish Toxins (PSTs) in Toxic Alexandrium catenella is Intertwined with Photosynthesis and Energy Production. Toxins. 2020; 12(8):477. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12080477
Chicago/Turabian StyleTse, Sirius Pui-kam, Fred Wang-fat Lee, Daniel Yun-lam Mak, Hang-kin Kong, Kenrick Kai-yuen Chan, Pak-yeung Lo, and Samuel Chun-lap Lo. 2020. "Production of Paralytic Shellfish Toxins (PSTs) in Toxic Alexandrium catenella is Intertwined with Photosynthesis and Energy Production" Toxins 12, no. 8: 477. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12080477





