OMICs Approaches in Diarrhetic Shellfish Toxins Research
Abstract
:1. Introduction
2. OMICs Overview
3. Human Toxicology of DSTs
4. Ecotoxicological Effects of DSTs
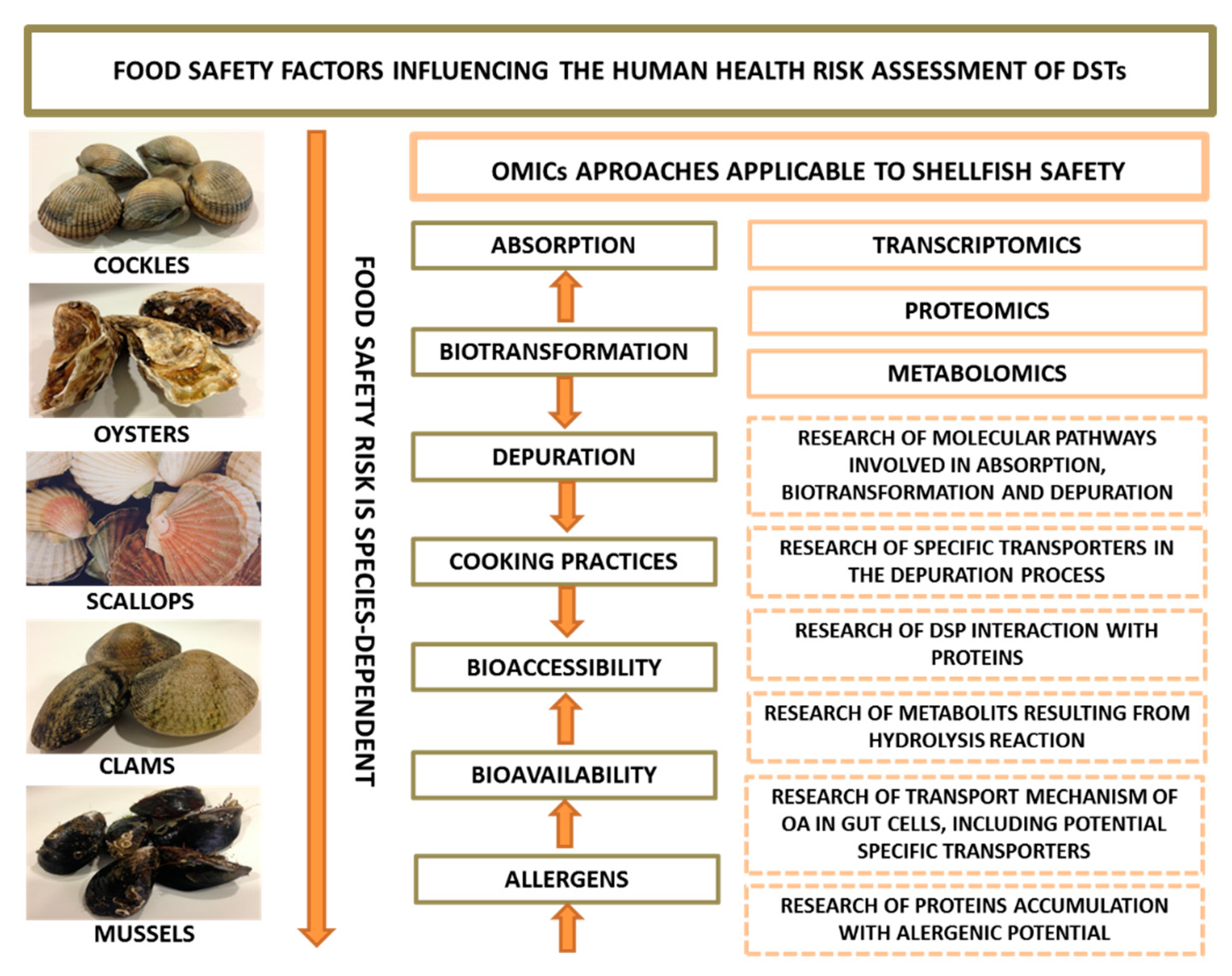
5. Food Safety
6. Shellfish Metabolism/Biotransformation of DSTs
7. Biomarkers of Exposure to DSTs
8. Concluding Remarks
- Further understanding of the molecular mechanisms associated with the toxicity of DSTs;
- Knowledge of the health effects associated to the chronic exposure of seafood consumers;
- Elucidating the mechanisms of transformation of DSTs in seafood, particularly in shellfish. There is an evident absence of molecular data concerning the bioactivity of the different DTX toxins and OA derivatives (esterified forms) relevant to the assessment of DSTs toxicity. There are other specific questions concerning the metabolic transformation of DSTs that can be elucidated employing metabolomics methods (e.g., chemical profile of FA-derivatives of OA and DTXs);
- OMICs will have a key role on the understanding the molecular processes that characterize bioavailability and bioaccessibility of DSTs, which is critical to promote an accurate risk assessment;
- Finally, proteomics can be particularly important in the investigation on the expression and accumulation of allergenic proteins in contaminated seafood and likely in assessing associations of DSTs with the protein matrix in contaminated seafood;
- Notwithstanding, OMICs breakthroughs in DSTs knowledge will be dependent on the improvement of the capacity of analysis of OMICs technologies. For instance, the access to thorough and well annotated genomic databases, particularly from marine invertebrates, will facilitate extracting biological information from complex gene and protein datasets. In proteomics, specific strategies, namely concerning protein fractionation, enrichment and isolation, are needed for capturing specific information about the metabolic pathways involved in DSTs toxicity and biotransformation.
Funding
Conflicts of Interest
References
- Visciano, P.; Schirone, M.; Berti, M.; Milandri, A.; Tofalo, R.; Suzzi, G. Marine Biotoxins: Occurrence, Toxicity, Regulatory Limits and Reference Methods. Front. Microbiol. 2016, 7, 1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vale, P.; Botelho, M.J.; Rodrigues, S.M.; Gomes, S.S.; Sampayo, M.A.D.M.; Henriques, M.J.B. Two decades of marine biotoxin monitoring in bivalves from Portugal (1986–2006): A review of exposure assessment. Harmful Algae 2008, 7, 11–25. [Google Scholar] [CrossRef]
- Reguera, B.; Riobó, P.; Rodríguez, F.; Díaz, P.A.; Pizarro, G.; Paz, B.; Franco, J.M.; Blanco, J. Dinophysis Toxins: Causative Organisms, Distribution and Fate in Shellfish. Mar. Drugs 2014, 12, 394–461. [Google Scholar] [CrossRef] [PubMed]
- Bauder, A.; Cembella, A.; Bricelj, V.; Quilliam, M. Uptake and fate of diarrhetic shellfish poisoning toxins from the dinoflagellate Prorocentrum lima in the bay scallop Argopecten irradians. Mar. Ecol. Prog. Ser. 2001, 213, 39–52. [Google Scholar] [CrossRef] [Green Version]
- Suganuma, M.; Fujiki, H.; Okabe, S.; Nishiwaki, S.; Brautigan, D.; Ingebritsen, T.S.; Rosner, M.R. Structurally different members of the okadaic acid class selectively inhibit protein serine/threonine but not tyrosine phosphatase activity. Toxicon 1992, 30, 873–878. [Google Scholar] [CrossRef]
- Shumway, S.E. Phycotoxin-related shellfish poisoning: Bivalve molluscs are not the only vectors. Rev. Fish. Sci. 1995, 3, 1–31. [Google Scholar] [CrossRef]
- The Occurrence of Prorocentrum Species [Algae] and Coincidental Gastrointestinal Illness of Mussel Consumers, Netherlands. Available online: https://agris.fao.org/agris-search/search.do?recordID=US19810650121 (accessed on 5 June 2020).
- Chan, L.L.; Hodgkiss, I.J.; Wan, J.M.-F.; Lum, J.H.-K.; Mak, A.S.-C.; Sit, W.-H.; Lo, S.C.L. Proteomic study of a model causative agent of harmful algal blooms, Prorocentrum triestinum II: The use of differentially expressed protein profiles under different growth phases and growth conditions for bloom prediction. Proteomics 2004, 4, 3214–3226. [Google Scholar] [CrossRef]
- Long, M.; Zhao, J.; Li, T.; Tafalla, C.; Zhang, Q.; Wang, X.; Gong, X.; Shen, Z.; Li, A. Transcriptomic and proteomic analyses of splenic immune mechanisms of rainbow trout (Oncorhynchus mykiss) infected by Aeromonas salmonicida subsp. salmonicida. J. Proteom. 2015, 122, 41–54. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, J.; He, Y.; Xie, Z.; Zhang, S.; Zhang, Y.; Lin, L.; Liu, S.; Wang, D.-Z. Quantitative proteomics reveals the key molecular events occurring at different cell cycle phases of the in situ blooming dinoflagellate cells. Sci. Total. Environ. 2019, 676, 62–71. [Google Scholar] [CrossRef]
- Zhang, S.-F.; Yuan, C.-J.; Chen, Y.; Lin, L.; Wang, D.-Z. Transcriptomic response to changing ambient phosphorus in the marine dinoflagellate Prorocentrum donghaiense. Sci. Total. Environ. 2019, 692, 1037–1047. [Google Scholar] [CrossRef]
- Swan, S.; Turner, A.D.; Bresnan, E.; Whyte, C.; Paterson, R.F.; McNeill, S.; Mitchell, E.; Davidson, K. Dinophysis acuta in Scottish Coastal Waters and Its Influence on Diarrhetic Shellfish Toxin Profiles. Toxins 2018, 10, 399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchida, H.; Watanabe, R.; Matsushima, R.; Oikawa, H.; Nagai, S.; Kamiyama, T.; Baba, K.; Miyazono, A.; Kosaka, Y.; Kaga, S.; et al. Toxin Profiles of Okadaic Acid Analogues and Other Lipophilic Toxins in Dinophysis from Japanese Coastal Waters. Toxins 2018, 10, 457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molinero-Abad, B.; Perez, L.; Izquierdo, D.; Escudero, I.; Arcos-Martínez, J. Sensor system based on flexible screen-printed electrodes for electrochemical detection of okadaic acid in seawater. Talanta 2019, 192, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Vilarinño, N.; Fonfría, E.S.; Louzao, M.C.; Botana, L.M. Use of Biosensors as Alternatives to Current Regulatory Methods for Marine Biotoxins. Sensors 2009, 9, 9414–9443. [Google Scholar] [CrossRef] [Green Version]
- Manita, D.; Alves, R.N.; Braga, A.; Fogaça, F.H.D.S.; Marques, A.; Costa, P.R. In vitro bioaccessibility of the marine biotoxins okadaic acid, dinophysistoxin-2 and their 7-O-acyl fatty acid ester derivatives in raw and steamed shellfish. Food Chem. Toxicol. 2017, 101, 121–127. [Google Scholar] [CrossRef]
- Alves, R.N.; Rambla-Alegre, M.; Braga, A.C.; Maulvault, A.L.; Barbosa, V.; Campàs, M.; Reverté, L.; Flores, C.; Caixach, J.; Kilcoyne, J.; et al. Bioaccessibility of lipophilic and hydrophilic marine biotoxins in seafood: An in vitro digestion approach. Food Chem. Toxicol. 2019, 129, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Kamat, P.K.; Rai, S.; Swarnkar, S.; Shukla, R.; Nath, C. Molecular and Cellular Mechanism of Okadaic Acid (OKA)-Induced Neurotoxicity: A Novel Tool for Alzheimer’s Disease Therapeutic Application. Mol. Neurobiol. 2014, 50, 852–865. [Google Scholar] [CrossRef]
- Rubiolo, J.; Lopez-Alonso, H.; Vega, F.; Vieytes, M.; Botana, L.M. Comparative study of toxicological and cell cycle effects of okadaic acid and dinophysistoxin-2 in primary rat hepatocytes. Life Sci. 2012, 90, 416–423. [Google Scholar] [CrossRef]
- Jiménez, D.; García, C.; Contreras, H.R. Toxins of Okadaic Acid-Group Increase Malignant Properties in Cells of Colon Cancer. Toxins 2020, 12, 179. [Google Scholar] [CrossRef] [Green Version]
- Fujiki, H.; Sueoka, E.; Watanabe, T.; Suganuma, M. The concept of the okadaic acid class of tumor promoters is revived in endogenous protein inhibitors of protein phosphatase 2A, SET and CIP2A, in human cancers. J. Cancer Res. Clin. Oncol. 2018, 144, 2339–2349. [Google Scholar] [CrossRef] [Green Version]
- Larsen, K.; Petersen, D.; Wilkins, A.L.; Samdal, I.; Sandvik, M.; Rundberget, T.; Goldstone, D.C.; Arcus, V.; Hovgaard, P.; Rise, F.; et al. Clarification of the C-35 Stereochemistries of Dinophysistoxin-1 and Dinophysistoxin-2 and Its Consequences for Binding to Protein Phosphatase. Chem. Res. Toxicol. 2007, 20, 868–875. [Google Scholar] [CrossRef]
- Ju, Z.; Wells, M.C.; Walter, R.B. DNA microarray technology in toxicogenomics of aquatic models: Methods and applications. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2007, 145, 5–14. [Google Scholar] [CrossRef]
- Gomes, F.; Watanabe, L.; Vianez, J.; Nunes, M.; Cardoso, J.; Lima, C.; Schneider, H.; Sampaio, I. Comparative analysis of the transcriptome of the Amazonian fish species Colossoma macropomum (tambaqui) and hybrid tambacu by next generation sequencing. PLoS ONE 2019, 14, e0212755. [Google Scholar] [CrossRef]
- Chen, W.; He, S. Isolation and characterization of microsatellite markers in a highland fish, Pareuchiloglanis sinensis (Siluriformes: Sisoridae) by next-generation sequencing. J. Genet. 2018, 97, 111–116. [Google Scholar] [CrossRef]
- Hill, J.J.; Callaghan, D.A.; Ding, W.; Kelly, J.F.; Chakravarthy, B.R. Identification of okadaic acid-induced phosphorylation events by a mass spectrometry approach. Biochem. Biophys. Res. Commun. 2006, 342, 791–799. [Google Scholar] [CrossRef]
- de Almeida, A.M.; Miller, I.; Eckersall, D. Proteomics in Domestic Animals: From Farm to Systems Biology; Springer International Publishing: Berlin/Heidelberg, Germany, 2018; ISBN 9783319696829. [Google Scholar]
- Huang, L.; Zou, Y.; Weng, H.-W.; Li, H.; Liu, J.; Yang, W. Proteomic profile in Perna viridis after exposed to Prorocentrum lima, a dinoflagellate producing DSP toxins. Environ. Pollut. 2015, 196, 350–357. [Google Scholar] [CrossRef]
- Cheung, C.H.Y.; Juan, H.-F. Quantitative proteomics in lung cancer. J. Biomed. Sci. 2017, 24, 37. [Google Scholar] [CrossRef]
- Paredi, G.; Sentandreu, M.-A.; Mozzarelli, A.; Fadda, S.; Hollung, K.; Almeida, A.M. Muscle and meat: New horizons and applications for proteomics on a farm to fork perspective. J. Proteom. 2013, 88, 58–82. [Google Scholar] [CrossRef]
- Domínguez-Pérez, D.; Lippolis, J.; Dennis, M.; Miller, B.; Tiley, K.; Vasconcelos, V.; De Almeida, A.M.; Campos, A. The Queen Conch (Lobatus gigas) Proteome: A Valuable Tool for Biological Studies in Marine Gastropods. Protein J. 2019, 38, 628–639. [Google Scholar] [CrossRef]
- Azevedo, C.C.; Guzmán-Guillén, R.; Martins, J.; Osório, H.; Vasconcelos, V.; Da Fonseca, R.R.; Campos, A. Proteomic profiling of gill GSTs in Mytilus galloprovincialis from the North of Portugal and Galicia evidences variations at protein isoform level with a possible relation with water quality. Mar. Environ. Res. 2015, 110, 152–161. [Google Scholar] [CrossRef]
- Almeida, A.M.; Bassols, A.; Bendixen, E.; Bhide, M.; Ceciliani, F.; Cristóbal, S.; E Silva, F.C.; Hollung, K.; Lisacek, F.; Mazzucchelli, G.; et al. Animal board invited review: Advances in proteomics for animal and food sciences. Animal 2014, 9, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Marco-Ramell, A.; Almeida, A.M.; Cristóbal, S.; Rodrigues, P.M.; Roncada, P.; Bassols, A. Proteomics and the search for welfare and stress biomarkers in animal production in the one-health context. Mol. BioSyst. 2016, 12, 2024–2035. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.; Almeida, A.M. Top-Down Proteomics and Farm Animal and Aquatic Sciences. Proteomes 2016, 4, 38. [Google Scholar] [CrossRef] [Green Version]
- Soares, R.; Franco, C.F.; Pires, E.; Ventosa, M.; Palhinhas, R.; Kočí, K.; Almeida, A.M.; Coelho, A.V. Mass spectrometry and animal science: Protein identification strategies and particularities of farm animal species. J. Proteom. 2012, 75, 4190–4206. [Google Scholar] [CrossRef]
- Rodrigues, P.M.; Schrama, D.; Campos, A.; Osorio, H.; Freitas, M. Applications of Proteomics in Aquaculture; Springer: Cham, Switzerland, 2016; ISBN 9783319432755. [Google Scholar]
- Murgarella, M.; Puiu, D.; Novoa, B.; Figueras, A.; Posada, D.; Canchaya, C. A first insight into the genome of the filter-feeder mussel Mytilus galloprovincialis. PLoS ONE 2016, 11, e0151561. [Google Scholar]
- Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.-L.; Wang, X.; Qi, H.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef] [Green Version]
- Badreddine, S.; Khazri, A.; Louati, H.; Dellali, M.; Driss, M.R.; Aïssa, P.; Mahmoudi, E.; Hamouda, B.; Coelho, A.V.; Sheehan, D. Effects of anthracene on filtration rates, antioxidant defense system, and redox proteomics in the Mediterranean clam Ruditapes decussatus (Mollusca: Bivalvia). Environ. Sci. Pollut. Res. 2015, 22, 10956–10968. [Google Scholar] [CrossRef]
- Romero, M.R.; Pérez-Figueroa, A.; Carrera, M.; Swanson, W.J.; Skibinski, D.O.; Diz, Á.P. RNA-seq coupled to proteomic analysis reveals high sperm proteome variation between two closely related marine mussel species. J. Proteom. 2019, 192, 169–187. [Google Scholar] [CrossRef]
- Zmasek, C.; Godzik, A. This Déjà Vu Feeling—Analysis of Multidomain Protein Evolution in Eukaryotic Genomes. PLoS Comput. Boil. 2012, 8, e1002701. [Google Scholar] [CrossRef] [Green Version]
- Petrak, J.; Ivanek, R.; Toman, O.; Cmejla, R.; Cmejlova, J.; Vyoral, D.; Živný, J.; Vulpe, C.D. Déjà vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics 2008, 8, 1744–1749. [Google Scholar] [CrossRef]
- Carpentier, S.; Panis, B.; Swennen, R.; Lammertyn, J. Finding the significant markers: Statistical analysis of proteomic data. Methods Mol. Boil. (Clifton N.J.) 2008, 428, 327–347. [Google Scholar]
- Fadda, S.; Almeida, A. Proteomics in Argentina-limitations and future perspectives: A special emphasis on meat proteomics. Proteomics 2015, 15, 3676–3687. [Google Scholar] [CrossRef]
- Bialojan, C.; Takai, A. Inhibitory effect of a marine sponge toxin, okadaic acid, on protein phosphatases. Biochem. J. 1988, 256, 283–290. [Google Scholar] [CrossRef]
- Oliveira, J.; Silva, C.B.D.C.E.; Mueller, T.; Martins, T.; Cova, M.; Silva, O.A.B.D.C.E.; Henriques, A.G. Toward Neuroproteomics in Biological Psychiatry: A Systems Approach Unravels Okadaic Acid-Induced Alterations in the Neuronal Phosphoproteome. OMICS: A J. Integr. Boil. 2017, 21, 550–563. [Google Scholar] [CrossRef]
- Yadav, L.; Tamene, F.; Göös, H.; Van Drogen, A.; Katainen, R.; Aebersold, R.; Gstaiger, M.; Varjosalo, M. Systematic Analysis of Human Protein Phosphatase Interactions and Dynamics. Cell Syst. 2017, 4, 430–444.e5. [Google Scholar] [CrossRef] [Green Version]
- Xing, Y.; Xu, Y.; Chen, Y.; Jeffrey, P.D.; Chao, Y.; Lin, Z.; Li, Z.; Strack, S.; Stock, J.B.; Shi, Y. Structure of Protein Phosphatase 2A Core Enzyme Bound to Tumor-Inducing Toxins. Cell 2006, 127, 341–353. [Google Scholar] [CrossRef] [Green Version]
- Maynes, J.T.; Bateman, K.S.; Cherney, M.M.; Das, A.K.; Luu, H.A.; Holmes, C.F.B.; James, M.N.G. Crystal Structure of the Tumor-promoter Okadaic Acid Bound to Protein Phosphatase-1. J. Boil. Chem. 2001, 276, 44078–44082. [Google Scholar] [CrossRef] [Green Version]
- Suganuma, M.; Fujiki, H.; Suguri, H.; Yoshizawa, S.; Hirota, M.; Nakayasu, M.; Ojika, M.; Wakamatsu, K.; Yamada, K.; Sugimura, T. Okadaic acid: An additional non-phorbol-12-tetradecanoate-13-acetate-type tumor promoter. Proc. Natl. Acad. Sci. USA 1988, 85, 1768–1771. [Google Scholar] [CrossRef] [Green Version]
- Opsahl, J.A.; Ljostveit, S.; Solstad, T.; Risa, K.; Roepstorff, P.; Fladmark, K.E. Identification of Dynamic Changes in Proteins Associated with the Cellular Cytoskeleton after Exposure to Okadaic Acid. Mar. Drugs 2013, 11, 1763–1782. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, Y.-Y.; Lin, L.; Gao, Y.; Hong, H.-S.; Wang, D.-Z. Quantitative proteomic analysis of okadaic acid treated mouse small intestines reveals differentially expressed proteins involved in diarrhetic shellfish poisoning. J. Proteom. 2012, 75, 2038–2052. [Google Scholar] [CrossRef]
- Hwang, J.; Pallas, D.C. STRIPAK complexes: Structure, biological function, and involvement in human diseases. Int. J. Biochem. Cell Boil. 2013, 47, 118–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merolla, F.; Luise, C.; Muller, M.T.; Pacelli, R.; Fusco, A.; Celetti, A. Loss of CCDC6, the First Identified RET Partner Gene, Affects pH2AX S139 Levels and Accelerates Mitotic Entry upon DNA Damage. PLoS ONE 2012, 7, e36177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirón-García, M.C.; Garrido-Godino, A.I.; Martínez-Fernández, V.; Fernández-Pevida, A.; Cuevas-Bermúdez, A.; Martín-Expósito, M.; Chávez, S.; De La Cruz, J.; Navarro, F. The yeast prefoldin-like URI-orthologue Bud27 associates with the RSC nucleosome remodeler and modulates transcription. Nucleic Acids Res. 2014, 42, 9666–9676. [Google Scholar] [CrossRef] [PubMed]
- Djouder, N.; Metzler, S.C.; Schmidt, A.; Wirbelauer, C.; Gstaiger, M.; Aebersold, R.; Hess, D.; Krek, W. S6K1-Mediated Disassembly of Mitochondrial URI/PP1γ Complexes Activates a Negative Feedback Program that Counters S6K1 Survival Signaling. Mol. Cell 2007, 28, 28–40. [Google Scholar] [CrossRef]
- Sala, G.L.; Ronzitti, G.; Sasaki, M.; Fuwa, H.; Yasumoto, T.; Bigiani, A.; Rossini, G.P. Proteomic Analysis Reveals Multiple Patterns of Response in Cells Exposed to a Toxin Mixture. Chem. Res. Toxicol. 2009, 22, 1077–1085. [Google Scholar] [CrossRef]
- Bodero, M.; Hoogenboom, L.; Bovee, T.; Portier, L.; De Haan, L.; Peijnenburg, A.; Hendriksen, P.J. Whole genome mRNA transcriptomics analysis reveals different modes of action of the diarrheic shellfish poisons okadaic acid and dinophysis toxin-1 versus azaspiracid-1 in Caco-2 cells. Toxicol. Vitr. 2018, 46, 102–112. [Google Scholar] [CrossRef]
- Dietrich, J.; Sommersdorf, C.; Gohlke, S.; Poetz, O.; Traenkle, B.; Rothbauer, U.; Hessel, S.; Lampen, A.; Braeuning, A. Okadaic acid activates Wnt/β-catenin-signaling in human HepaRG cells. Arch. Toxicol. 2019, 93, 1927–1939. [Google Scholar] [CrossRef]
- Vieira, A.C.; Rubiolo, J.A.; López-Alonso, H.; Cifuentes, J.M.; Alfonso, A.; Bermúdez, R.; Otero, P.; Vieytes, M.R.; Vega, F.V.; Botana, L.M. Oral Toxicity of Okadaic Acid in Mice: Study of Lethality, Organ Damage, Distribution and Effects on Detoxifying Gene Expression. Toxins 2013, 5, 2093–2108. [Google Scholar] [CrossRef] [Green Version]
- Valdiglesias, V.; Fernandez-Tajes, J.; Méndez, J.; Pásaro, E.; Laffon, B. The marine toxin okadaic acid induces alterations in the expression level of cancer-related genes in human neuronal cells. Ecotoxicol. Environ. Saf. 2013, 92, 303–311. [Google Scholar] [CrossRef]
- Marine biotoxins in shellfish—okadaic acid and analogues-Scientific Opinion of the Panel on Contaminants in the Food chain. EFSA J. 2008, 6, 589. [CrossRef]
- Lopes, V.; Costa, P.; Rosa, R. Effects of Harmful Algal Bloom Toxins on Marine Organisms. In Ecotoxicology of Marine Organisms; CRC Press: Boca Raton, FL, USA, 2019; pp. 42–88. [Google Scholar]
- Blanco, J.; Marino, C.; Martin, H.; Acosta, C.P. Anatomical distribution of diarrhetic shellfish poisoning (DSP) toxins in the mussel Mytilus galloprovincialis. Toxicon 2007, 50, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- McCarron, P.; Kilcoyne, J.; Hess, P. Effects of cooking and heat treatment on concentration and tissue distribution of okadaic acid and dinophysistoxin-2 in mussels (Mytilus edulis). Toxicon 2008, 51, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- Blanco, J. Accumulation of Dinophysis Toxins in Bivalve Molluscs. Toxins 2018, 10, 453. [Google Scholar] [CrossRef] [Green Version]
- Romero-Geraldo, R.D.J.; García-Lagunas, N.; Hernández-Saavedra, N.Y. Effects of In Vitro Exposure to Diarrheic Toxin Producer Prorocentrum lima on Gene Expressions Related to Cell Cycle Regulation and Immune Response in Crassostrea gigas. PLoS ONE 2014, 9, e97181. [Google Scholar] [CrossRef]
- Romero-Geraldo, R.D.J.; García-Lagunas, N.; Hernández-Saavedra, N.Y. Crassostrea gigas exposure to the dinoflagellate Prorocentrum lima: Histological and gene expression effects on the digestive gland. Mar. Environ. Res. 2016, 120, 93–102. [Google Scholar] [CrossRef]
- Neves, R.A.; Santiago, T.C.; Carvalho, W.; Silva, E.D.S.; Da Silva, P.M.; Nascimento, S.M. Impacts of the toxic benthic dinoflagellate Prorocentrum lima on the brown mussel Perna perna: Shell-valve closure response, immunology, and histopathology. Mar. Environ. Res. 2019, 146, 35–45. [Google Scholar] [CrossRef]
- Nielsen, P.; Krock, B.; Hansen, P.J.; Vismann, B. Effects of the DSP-toxic dinoflagellate Dinophysis acuta on clearance and respiration rate of the blue mussel, Mytilus edulis. PLoS ONE 2020, 15, e0230176. [Google Scholar] [CrossRef] [Green Version]
- Prego-Faraldo, M.V.; Vieira, L.; Eirin-Lopez, J.; Méndez, J.; Guilhermino, L.; M?ndez, J. Transcriptional and biochemical analysis of antioxidant enzymes in the mussel Mytilus galloprovincialis during experimental exposures to the toxic dinoflagellate Prorocentrum lima. Mar. Environ. Res. 2017, 129, 304–315. [Google Scholar] [CrossRef]
- Wei, X.-M.; Lu, M.-Y.; Duan, G.-F.; Li, H.-Y.; Liu, J.-S.; Yang, W.-D. Responses of CYP450 in the mussel Perna viridis after short-term exposure to the DSP toxins-producing dinoflagellate Prorocentrum lima. Ecotoxicol. Environ. Saf. 2019, 176, 178–185. [Google Scholar] [CrossRef]
- Zou, Y.; Wei, X.-M.; Weng, H.-W.; Li, H.; Liu, J.; Yang, W. Expression profile of eight glutathione S-transferase genes in Crassostrea ariakensis after exposure to DSP toxins producing dinoflagellate Prorocentrum lima. Toxicon 2015, 105, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wang, J.; Chen, W.-C.; Li, H.; Liu, J.-S.; Jiang, T.; Yang, W.-D. P-glycoprotein expression in Perna viridis after exposure to Prorocentrum lima, a dinoflagellate producing DSP toxins. Fish Shellfish. Immunol. 2014, 39, 254–262. [Google Scholar] [CrossRef]
- Huang, L.; Liu, S.-L.; Zheng, J.-W.; Li, H.; Liu, J.; Yang, W. P-glycoprotein and its inducible expression in three bivalve species after exposure to Prorocentrum lima. Aquat. Toxicol. 2015, 169, 123–132. [Google Scholar] [CrossRef]
- Manfrin, C.; Dreos, R.; Battistella, S.; Beran, A.; Gerdol, M.; Varotto, L.; Lanfranchi, G.; Venier, P.; Pallavicini, A. Mediterranean Mussel Gene Expression Profile Induced by Okadaic Acid Exposure. Environ. Sci. Technol. 2010, 44, 8276–8283. [Google Scholar] [CrossRef]
- Chi, C.; Giri, S.S.; Jun, J.W.; Kim, S.W.; Kim, H.J.; Kang, J.W.; Park, S.C. Detoxification- and Immune-Related Transcriptomic Analysis of Gills from Bay Scallops (Argopectenirradians) in Response to Algal Toxin Okadaic Acid. Toxins 2018, 10, 308. [Google Scholar] [CrossRef] [Green Version]
- Bánfi, B.; Malgrange, B.; Knisz, J.; Steger, K.; Dubois-Dauphin, M.; Krause, K.-H. NOX3, a Superoxide-generating NADPH Oxidase of the Inner Ear. J. Boil. Chem. 2004, 279, 46065–46072. [Google Scholar] [CrossRef] [Green Version]
- Madamanchi, N.R.; Li, S.; Patterson, C.; Runge, M.S. Reactive oxygen species regulate heat-shock protein 70 via the JAK/STAT pathway. Arter. Thromb. Vasc. Boil. 2001, 21, 321–326. [Google Scholar] [CrossRef] [Green Version]
- Suarez-Ulloa, V.; Fernandez-Tajes, J.; Aguiar-Pulido, V.; Prego-Faraldo, M.V.; Florez-Barros, F.; Sexto-Iglesias, A.; Mendez, J.; Eirin-Lopez, J.M. Unbiased high-throughput characterization ofmussel transcriptomic responses to sublethal concentrations of the biotoxin okadaic acid. PeerJ 2015, 2015, e1429. [Google Scholar] [CrossRef] [Green Version]
- Prego-Faraldo, M.V.; Martínez, L.; Mendez, J. RNA-Seq Analysis for Assessing the Early Response to DSP Toxins in Mytilus galloprovincialis Digestive Gland and Gill. Toxins 2018, 10, 417. [Google Scholar] [CrossRef] [Green Version]
- Miao, J.; Chi, L.; Pan, L.; Song, Y. Generally detected genes in comparative transcriptomics in bivalves: Toward the identification of molecular markers of cellular stress response. Environ. Toxicol. Pharmacol. 2015, 39, 475–481. [Google Scholar] [CrossRef]
- Dou, M.; Jiao, Y.; Zheng, J.; Zhang, G.; Li, H.; Liu, J.; Yang, W. De novo transcriptome analysis of the mussel Perna viridis after exposure to the toxic dinoflagellate Prorocentrum lima. Ecotoxicol. Environ. Saf. 2020, 192, 110265. [Google Scholar] [CrossRef] [PubMed]
- Le Du, J.; Tovar-Ramírez, D.; Núñez-Vázquez, E. Embryotoxic effects of dissolved okadaic acid on the development of Longfin yellowtail Seriola rivoliana. Aquat. Toxicol. 2017, 190, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Escoffier, N.; Gaudin, J.; Mezhoud, K.; Huet, H.; Château-Joubert, S.; Turquet, J.; Crespeau, F.; Edery, M. Toxicity to medaka fish embryo development of okadaic acid and crude extracts of Prorocentrum dinoflagellates. Toxicon 2007, 49, 1182–1192. [Google Scholar] [CrossRef]
- Ajuzie, C.C. Toxic Prorocentrum lima induces abnormal behaviour in juvenile sea bass. Environ. Boil. Fishes 2007, 20, 19–27. [Google Scholar] [CrossRef]
- Souid, G.; Souayed, N.; Haouas, Z.; Maaroufi, K. Does the phycotoxin Okadaic acid cause oxidative stress damages and histological alterations to seabream (Sparus aurata)? Toxicon 2018, 144, 55–60. [Google Scholar] [CrossRef]
- Corriere, M.; Baptista, M.; Paula, J.R.; Repolho, T.; Rosa, R.; Costa, P.R.; Soliño, L. Impaired fish swimming performance following dietary exposure to the marine phycotoxin okadaic acid. Toxicon 2020, 179, 53–59. [Google Scholar] [CrossRef]
- Zhang, N.S.; Li, H.Y.; Liu, J.S.; Yang, W.D. Gene expression profiles in zebrafish (Danio rerio) liver after acute exposure to okadaic acid. Environ. Toxicol. Pharmacol. 2014, 37, 791–802. [Google Scholar] [CrossRef]
- Yasumoto, T.; Murata, M.; Oshima, Y.; Sano, M.; Matsumoto, G.; Clardy, J. Diarrhetic shellfish toxins. Tetrahedron 1985, 41, 1019–1025. [Google Scholar] [CrossRef]
- Costa, P.R.; Costa, S.T.; Braga, A.; Rodrigues, S.M.; Vale, P. Relevance and challenges in monitoring marine biotoxins in non-bivalve vectors. Food Control. 2017, 76, 24–33. [Google Scholar] [CrossRef]
- Murk, A.J.; Nicolas, J.; Smulders, F.J.; Bürk, C.; Gerssen, A. Marine biotoxins: Types of poisoning, underlying mechanisms of action and risk management programmes. In ECVPH Food Safety Assurance; Wageningen Academic: Wageningen, Netherlands, 2019; pp. 207–239. [Google Scholar] [CrossRef]
- Vilarinño, N.; Louzao, M.C.; Abal, P.; Cagide, E.; Carrera, C.; Vieytes, M.R.; Botana, L.M. Human Poisoning from Marine Toxins: Unknowns for Optimal Consumer Protection. Toxins 2018, 10, 324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Ota, H.; Yamasaki, M. Direct evidence of transformation of dinophysistoxin-1 to 7-O-acyl-dinophysistoxin-1 (dinophysistoxin-3) in the scallop Patinopecten yessoensis. Toxicon 1999, 37, 187–198. [Google Scholar] [CrossRef]
- Takai, A.; Murata, M.; Isobe, M.; Mieskes, G.; Yasumoto, T. Inhibitory effect of okadaic acid derivatives on protein phosphatases. A study on structure-affinity relationship. Biochem. J. 1992, 284, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Vale, P. Differential dynamics of dinophysistoxins and pectenotoxins between blue mussel and common cockle: A phenomenon originating from the complex toxin profile of Dinophysis acuta. Toxicon 2004, 44, 123–134. [Google Scholar] [CrossRef]
- Rossignoli, A.E.; Fernandez, D.; Regueiro, J.; Marino, C.; Blanco, J. Esterification of okadaic acid in the mussel Mytilus galloprovincialis. Toxicon 2011, 57, 712–720. [Google Scholar] [CrossRef]
- Vale, P.; Sampayo, M.A.D.M. First confirmation of human diarrhoeic poisonings by okadaic acid esters after ingestion of razor clams (Solen marginatus) and green crabs (Carcinus maenas) in Aveiro lagoon, Portugal and detection of okadaic acid esters in phytoplankton. Toxicon 2002, 40, 989–996. [Google Scholar] [CrossRef]
- Suzuki, T.; Mitsuya, T. Comparison of dinophysistoxin-1 and esterified dinophysistoxin-1 (dinophysistoxin-3) contents in the scallop Patinopecten yessoensis and the mussel Mytilus galloprovincialis. Toxicon 2001, 39, 905–908. [Google Scholar] [CrossRef]
- Nzoughet, J.K.; Hamilton, J.T.G.; Botting, C.H.; Douglas, A.; Devine, L.; Nelson, J.; Elliott, C.T. Proteomics Identification of Azaspiracid Toxin Biomarkers in Blue Mussels, Mytilus edulis. Mol. Cell. Proteom. 2009, 8, 1811–1822. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.-P.H.; Yap, E.-H.; Xiao, H.; Fiser, A.; Horwitz, S.B. 2-(m-Azidobenzoyl)taxol binds differentially to distinct β-tubulin isotypes. Proc. Natl. Acad. Sci. USA 2016, 113, 11294–11299. [Google Scholar] [CrossRef] [Green Version]
- Cui, H.H.T. Uncovering Drug Mechanism of Action by Proteome Wide- Identification of Drug-Binding Proteins. Med. Chem. 2017, 13, 13. [Google Scholar] [CrossRef]
- Shin, N.-Y.; Liu, Q.; Stamer, A.S.L.; Liebler, D.C. Protein Targets of Reactive Electrophiles in Human Liver Microsomes. Chem. Res. Toxicol. 2007, 20, 859–867. [Google Scholar] [CrossRef] [Green Version]
- Carlsson, H.; Törnqvist, M. Strategy for identifying unknown hemoglobin adducts using adductome LC-MS/MS data: Identification of adducts corresponding to acrylic acid, glyoxal, methylglyoxal, and 1-octen-3-one. Food Chem. Toxicol. 2016, 92, 94–103. [Google Scholar] [CrossRef] [PubMed]
- EUR-Lex-32004R0853-EN-EUR-Lex. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32004R0853 (accessed on 19 June 2020).
- Picot, C.; Limon, G.; Durand, G.; Wesolek, N.; Parent-Massin, D.; Roudot, A.-C. Domoic Acid, Okadaic Acid and Spirolides: Inter-Species Variability in Contamination and Cooking Effects. Food Public Health 2012, 2, 50–57. [Google Scholar] [CrossRef] [Green Version]
- Blanco, J.; Arévalo, F.; Correa, J.; Porro, M.C.; Cabado, A.G.; Vieites, J.M.; Moroño, Á. Effect of the industrial steaming on the toxicity, estimated by LC–MS/MS, of mussels exposed for a long time to diarrhetic shellfish poisoning (DSP) toxins. Food Chem. 2015, 177, 240–247. [Google Scholar] [CrossRef] [PubMed]
- García, C.; Oyaneder-Terrazas, J.; Contreras, C.; Del Campo, M.; Torres, R.; Contreras, H.R. Determination of the toxic variability of lipophilic biotoxins in marine bivalve and gastropod tissues treated with an industrial canning process. Food Addit. Contam. Part A 2016, 33, 1711–1727. [Google Scholar] [CrossRef]
- Braga, A.; Alves, R.N.; Maulvault, A.L.; Barbosa, V.; Marques, A.; Costa, P.R. In vitro bioaccessibility of the marine biotoxin okadaic acid in shellfish. Food Chem. Toxicol. 2016, 89, 54–59. [Google Scholar] [CrossRef]
- EUR-Lex-32011R0015-EN-EUR-Lex. Available online: https://eur-lex.europa.eu/eli/reg/2011/15/oj (accessed on 19 June 2020).
- Ehlers, A.; Scholz, J.; These, A.; Hessel, S.; Preiss-Weigert, A.; Lampen, A. Analysis of the passage of the marine biotoxin okadaic acid through an in vitro human gut barrier. Toxicol. 2011, 279, 196–202. [Google Scholar] [CrossRef]
- Rahman, A.M.A.; Kamath, S.D.; Lopata, A.; Helleur, R.J. Analysis of the allergenic proteins in black tiger prawn (Penaeus monodon) and characterization of the major allergen tropomyosin using mass spectrometry. Rapid Commun. Mass Spectrom. 2010, 24, 2462–2470. [Google Scholar] [CrossRef]
- Ruethers, T.; Taki, A.C.; Johnston, E.; Nugraha, R.; Le, T.T.K.; Kalic, T.; McLean, T.; Kamath, S.D.; Lopata, A.L. Seafood allergy: A comprehensive review of fish and shellfish allergens. Mol. Immunol. 2018, 100, 28–57. [Google Scholar] [CrossRef]
- Manduzio, H.; Cosette, P.; Gricourt, L.; Jouenne, T.; Lenz, C.; Andersen, O.-K.; Leboulenger, F.; Rocher, B. Proteome modifications of blue mussel (Mytilus edulis L.) gills as an effect of water pollution. Proteomics 2005, 5, 4958–4963. [Google Scholar] [CrossRef]
- Vidal, A.; Ruiz, Y.; Suarez, P.; Martinez, A.A.; Rossignoli, A.E.; Blanco, J.; Garcia, O.; Juan, F.S. Accumulation of Okadaic Acid and Detoxifying Enzymes in the Digestive Gland of Mytilus galloprovincialis during Exposure to DSP. In Molluscan Shellfish Safety; Springer: Berlin/Heidelberg, Germany, 2013; pp. 217–225. [Google Scholar]
- Hu, T.; Curtis, J.M.; Walter, J.A.; Wright, J.L. Identification of DTX-4, a new water-soluble phosphatase inhibitor from the toxic dinoflagellate Prorocentrum lima. J. Chem. Soc., Chem. Commun. 1995, 597. [Google Scholar] [CrossRef]
- Cruz, P.G.; Daranas, A.H.; Fernández, J.J.; Souto, M.L.; Norte, M. DTX5c, a new OA sulphate ester derivative from cultures of Prorocentrum belizeanum. Toxicon 2006, 47, 920–924. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Leblanc, P.; Burton, I.W.; Walter, J.A.; McCarron, P.; Melanson, J.; Strangman, W.K.; Wright, J.L. Sulfated diesters of okadaic acid and DTX-1: Self-protective precursors of diarrhetic shellfish poisoning (DSP) toxins. Harmful Algae 2017, 63, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Curtis, J.M.; Walter, J.A.; McLachlan, J.L.; Wright, J.L. Two new water-soluble dsp toxin derivatives from the dinoflagellate prorocentrum maculosum: Possible storage and excretion products. Tetrahedron Lett. 1995, 36, 9273–9276. [Google Scholar] [CrossRef]
- Marr, J.; Hu, T.; Pleasance, S.; Quilliam, M.; Wright, J. Detection of new 7-O-acyl derivatives of diarrhetic shellfish poisoning toxins by liquid chromatography-mass spectrometry. Toxicon 1992, 30, 1621–1630. [Google Scholar] [CrossRef]
- Konoki, K.; Onoda, T.; Watanabe, R.; Cho, Y.; Kaga, S.; Suzuki, T.; Yotsu-Yamashita, M. In Vitro Acylation of Okadaic Acid in the Presence of Various Bivalves’ Extracts. Mar. Drugs 2013, 11, 300–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, R.S. Comparative genomics and proteomics of vertebrate diacylglycerol acyltransferase (DGAT), acyl CoA wax alcohol acyltransferase (AWAT) and monoacylglycerol acyltransferase (MGAT). Comp. Biochem. Physiol. Part D Genom. Proteom. 2010, 5, 45–54. [Google Scholar] [CrossRef] [Green Version]
- Torgersen, T.; Lindegarth, S.; Ungfors, A.; Sandvik, M. Profiles and levels of fatty acid esters of okadaic acid group toxins and pectenotoxins during toxin depuration, Part I: Brown crab (Cancer pagurus). Toxicon 2008, 52, 407–417. [Google Scholar] [CrossRef]
- Vale, P. Detailed profiles of 7-O-acyl esters in plankton and shellfish from the Portuguese coast. J. Chromatogr. A 2006, 1128, 181–188. [Google Scholar] [CrossRef]
- Blanco; Arévalo; Correa; Moroño Lipophilic Toxins in Galicia (NW Spain) between 2014 and 2017: Incidence on the Main Molluscan Species and Analysis of the Monitoring Efficiency. Toxins 2019, 11, 612. [CrossRef] [Green Version]
- Azpeitia, K.; Ferrer, L.; Revilla, M.; Pagaldai, J.; Mendiola, D. Growth, biochemical profile, and fatty acid composition of mussel (Mytilus galloprovincialis Lmk.) cultured in the open ocean of the Bay of Biscay (northern Spain). Aquaculture 2016, 454, 95–108. [Google Scholar] [CrossRef]
- Martinez, I.; Sánchez-Lazo, C.; Ruiz-Jarabo, I.; Herrera, M.; Mancera, J.M. Biochemical composition, lipid classes, fatty acids and sexual hormones in the mussel Mytilus galloprovincialis from cultivated populations in south Spain. Aquaculture 2012, 358, 274–283. [Google Scholar] [CrossRef]
- Gerssen, A.; Mulder, P.P.; De Boer, J. Screening of lipophilic marine toxins in shellfish and algae: Development of a library using liquid chromatography coupled to orbitrap mass spectrometry. Anal. Chim. Acta 2011, 685, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Torgersen, T.; Wilkins, A.L.; Rundberget, T.; Miles, C.O. Characterization of fatty acid esters of okadaic acid and related toxins in blue mussels (Mytilus edulis) from Norway. Rapid Commun. Mass Spectrom. 2008, 22, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- Contreras, H.R.; García, C. Inter-species variability of okadaic acid group toxicity in relation to the content of fatty acids detected in different marine vectors. Food Addit. Contam. Part A 2019, 36, 464–482. [Google Scholar] [CrossRef] [PubMed]
- Lindegarth, S.; Torgersen, T.; Lundve, B.; Sandvik, M. Differential Retention of Okadaic Acid (OA) Group Toxins and Pectenotoxins (PTX) in the Blue Mussel, Mytilus edulis(L.), and European Flat Oyster, Ostrea edulis(L.). J. Shellfish. Res. 2009, 28, 313–323. [Google Scholar] [CrossRef]
- De Oliveira, M.M.; Camanho, A.S.; Gaspar, M.B. The phycotoxins׳ impact on the revenue of the Portuguese artisanal dredge fleet. Mar. Policy 2015, 52, 45–51. [Google Scholar] [CrossRef]
- Liu, C.; Gu, C.; Huang, W.; Sheng, X.; Du, J.; Li, Y. Targeted UPLC-MS/MS high-throughput metabolomics approach to assess the purine and pyrimidine metabolism. J. Chromatogr. B 2019, 1113, 98–106. [Google Scholar] [CrossRef]
- Amacher, D.E. The discovery and development of proteomic safety biomarkers for the detection of drug-induced liver toxicity. Toxicol. Appl. Pharmacol. 2010, 245, 134–142. [Google Scholar] [CrossRef]
- Connon, R.E.; Geist, J.; Werner, I. Effect-Based Tools for Monitoring and Predicting the Ecotoxicological Effects of Chemicals in the Aquatic Environment. Sensors 2012, 12, 12741–12771. [Google Scholar] [CrossRef] [Green Version]
- Sant, G.R.; Knopf, K.B.; Albala, D.M. Live-single-cell phenotypic cancer biomarkers-future role in precision oncology? NPJ Precis. Oncol. 2017, 1, 1–7. [Google Scholar] [CrossRef] [Green Version]
- María, V.P.F. Assessment of the Early Effects of the Diarrhetic Shellfish Toxins in the Mussel Mytilus galloprovincialis Using Cellular and Molecular Biomarkers. Ph.D. Thesis, Universidade da Coruña, Coruña, Spain, 2016. [Google Scholar]
- Venier, P.; De Pitta, C.; Pallavicini, A.; Marsano, F.; Varotto, L.; Romualdi, C.; Dondero, F.; Viarengo, A.; Lanfranchi, G. Development of mussel mRNA profiling: Can gene expression trends reveal coastal water pollution? Mutat. Res. Mol. Mech. Mutagen. 2006, 602, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Chi, C.; Giri, S.S.; Jun, J.; Kim, H.; Kim, S.; Yun, S.; Park, S.C. Effects of algal toxin okadaic acid on the non-specific immune and antioxidant response of bay scallop (Argopecten irradians). Fish Shellfish. Immunol. 2017, 65, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Chi, C.; Giri, S.S.; Jun, J.W.; Kim, H.J.; Yun, S.; Kim, S.G.; Park, S.C. Marine Toxin Okadaic Acid Affects the Immune Function of Bay Scallop (Argopecten irradians). Molecules 2016, 21, 1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthews, K. Protein biomarker signatures for accurate diagnosis of urothelial cancer. Nat. Clin. Pr. Oncol. 2006, 3, 233. [Google Scholar] [CrossRef]
- He, Z.B.; Duan, G.F.; Liang, C.Y.; Li, H.Y.; Liu, J.S.; Yang, W.D. Up-regulation of Nrf2-dependent antioxidant defenses in Perna viridis after exposed to Prorocentrum lima. Fish Shellfish Immunol. 2019, 90, 173–179. [Google Scholar] [CrossRef]

| Functional Categories | Molecular Marker | Biological Function | References |
|---|---|---|---|
| Antioxidant system | Superoxide dismutase (SOD) | removal of superoxide radicals | [72] |
| Catalase (CAT) | hydrogen peroxide catabolic process | [72] | |
| Glutathione peroxidase (GPx, Se-GPx) | hydrogen peroxide catabolic process | [72] | |
| Glutathione S-Transferase (GST, GST-pi, GST-σ3, GST-ω) | glutathione peroxidase activity, xenobiotic catabolic process | [72,84,143] | |
| Metabolic detoxification | Cytochrome P450 (CYP2D14, CYP3A4, CYP3L3, CYP2C8, CYP3A) | xenobiotic metabolic process, oxidoreductase activity | [73] |
| ATP-binding cassette sub-family B member 10 (ABCB10) | transmembrane transport | [84] | |
| Multidrug resistance-associated protein (ABCC1, ABCG) | xenobiotic transmembrane transporter activity | [84,143] | |
| Transcription | Cyclic AMP-dependent transcription factor (ATF4-like) | regulation of transcription by RNA polymerase II | [84] |
| CREB/ATF bZIP transcription factor (CREBZF) | transcription regulation | [84] | |
| Nuclear factor erythroid 2-related factor (NRF2) | transcription factor that plays a key role in the response to oxidative stress | [84,143] | |
| Protein metabolism | Inhibitor of apoptosis protein (IAP) | ubiquitin-dependent protein catabolic process | [84] |
| Kelch-like ECH-associated protein 1 (KEAP1) | protein ubiquitination, cellular response to oxidative stress | [84,143] | |
| Cell structure | Tubulin alpha-1C chain (TUBA1C) | microtubule constituent | [84,143] |
| Tubulin beta-1 chain (TUBB1) | microtubule constituent | [84,143] | |
| Actin related protein (ARP 2, ARP 3) | mediates actin polymerization | [76] | |
| Immune response | annexin-like (ANXA) | regulator of the inflammatory process | [76] |
| Complement component 1q (mgC1q83 and mgC1q29) | pathogen recognition | [84] | |
| Regulation of metabolism and signal transduction | Heat shock protein HSP 90 (HSP90) | regulation of specific target proteins involved for instance in cell cycle control and signal transduction | [84] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos, A.; Freitas, M.; de Almeida, A.M.; Martins, J.C.; Domínguez-Pérez, D.; Osório, H.; Vasconcelos, V.; Reis Costa, P. OMICs Approaches in Diarrhetic Shellfish Toxins Research. Toxins 2020, 12, 493. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12080493
Campos A, Freitas M, de Almeida AM, Martins JC, Domínguez-Pérez D, Osório H, Vasconcelos V, Reis Costa P. OMICs Approaches in Diarrhetic Shellfish Toxins Research. Toxins. 2020; 12(8):493. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12080493
Chicago/Turabian StyleCampos, Alexandre, Marisa Freitas, André M. de Almeida, José Carlos Martins, Dany Domínguez-Pérez, Hugo Osório, Vitor Vasconcelos, and Pedro Reis Costa. 2020. "OMICs Approaches in Diarrhetic Shellfish Toxins Research" Toxins 12, no. 8: 493. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12080493






