Varying Protein Levels Influence Metabolomics and the Gut Microbiome in Healthy Adult Dogs
Abstract
:1. Introduction
2. Results
2.1. Proximate Analysis and Digestibility of Test Foods
2.2. Study Animals and Body Weight
2.3. Serum Metabolites
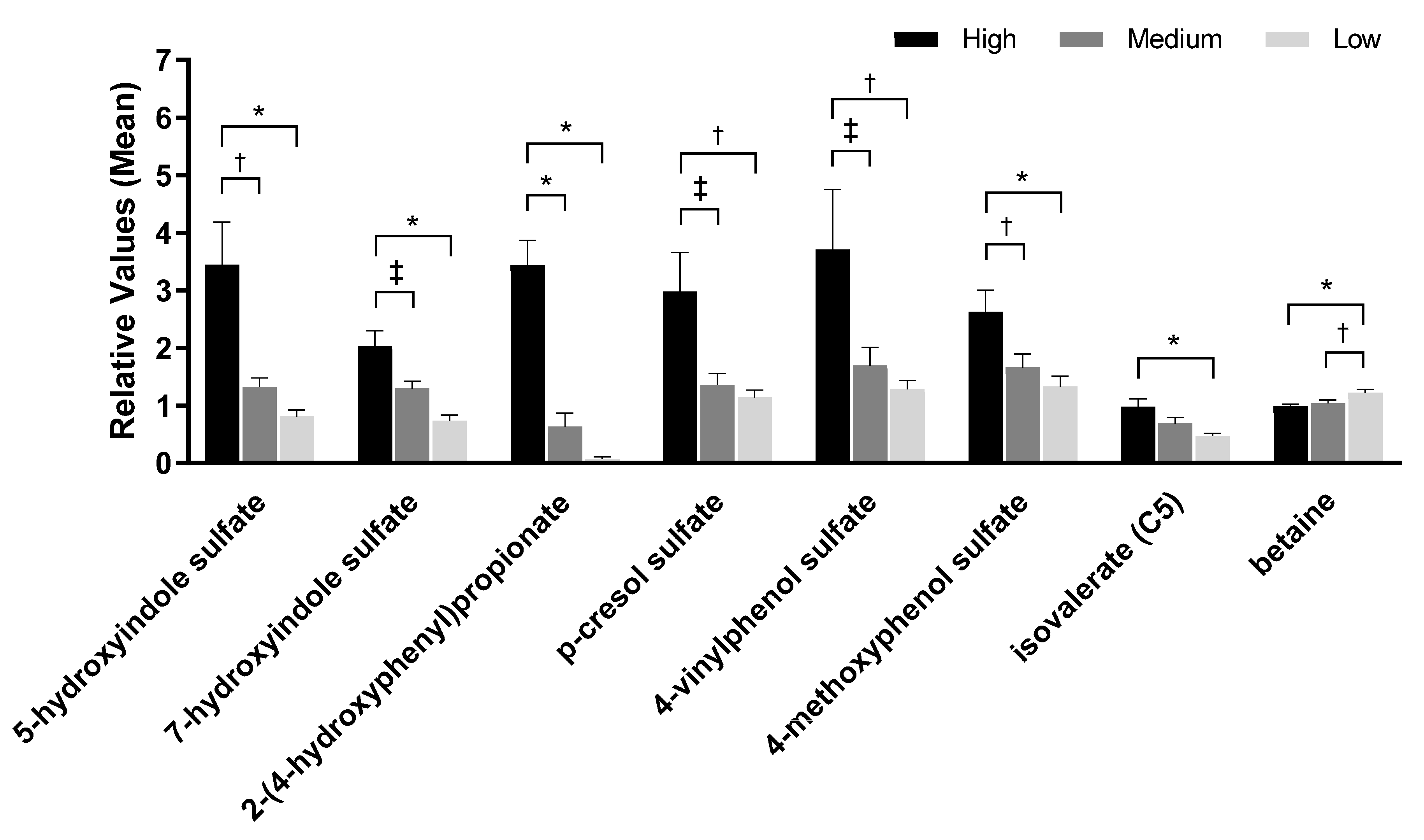
2.4. Urine Metabolites
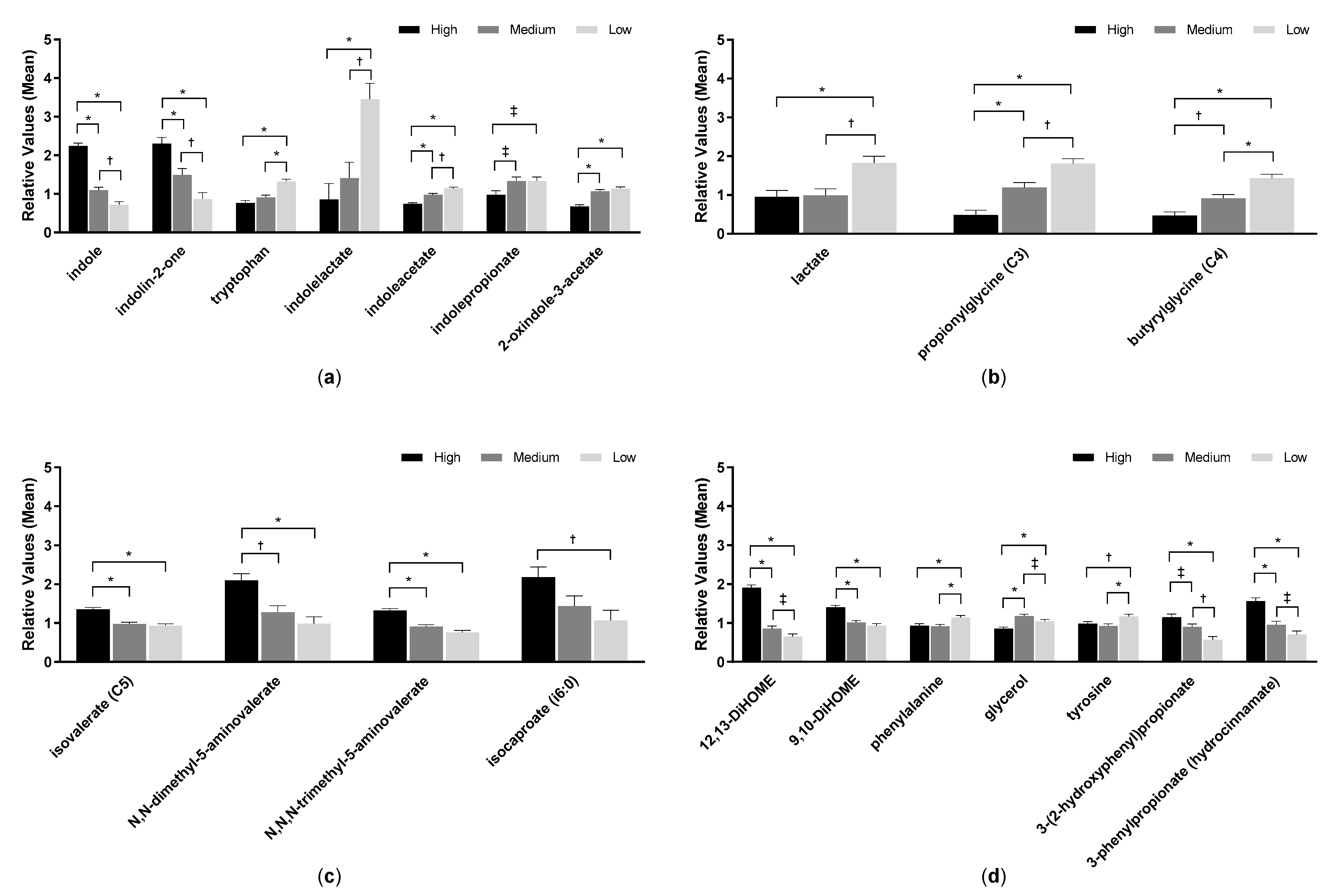
2.5. Fecal pH and Metabolites
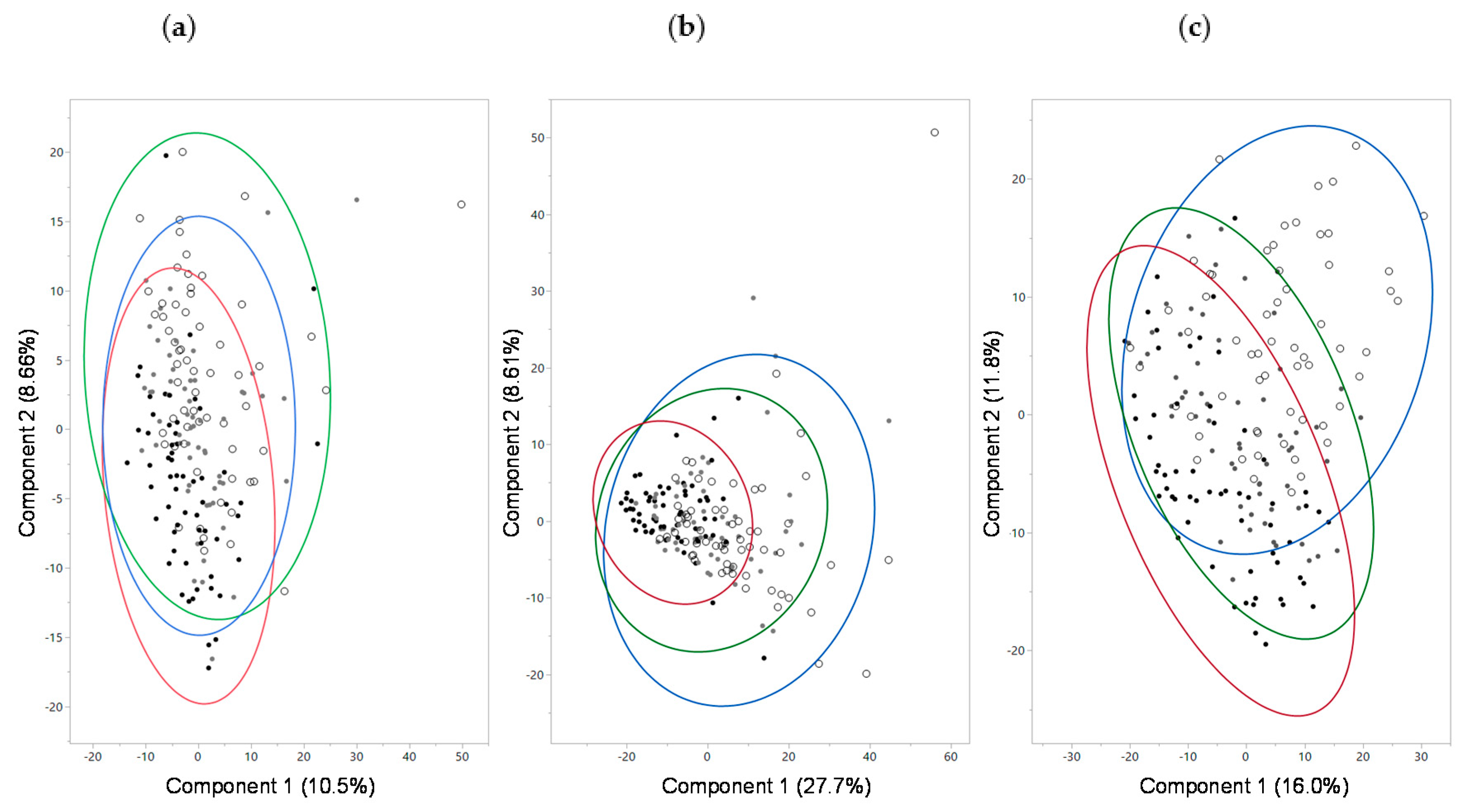
2.6. Fecal Microbiome
3. Discussion
4. Materials and Methods
4.1. Study Foods
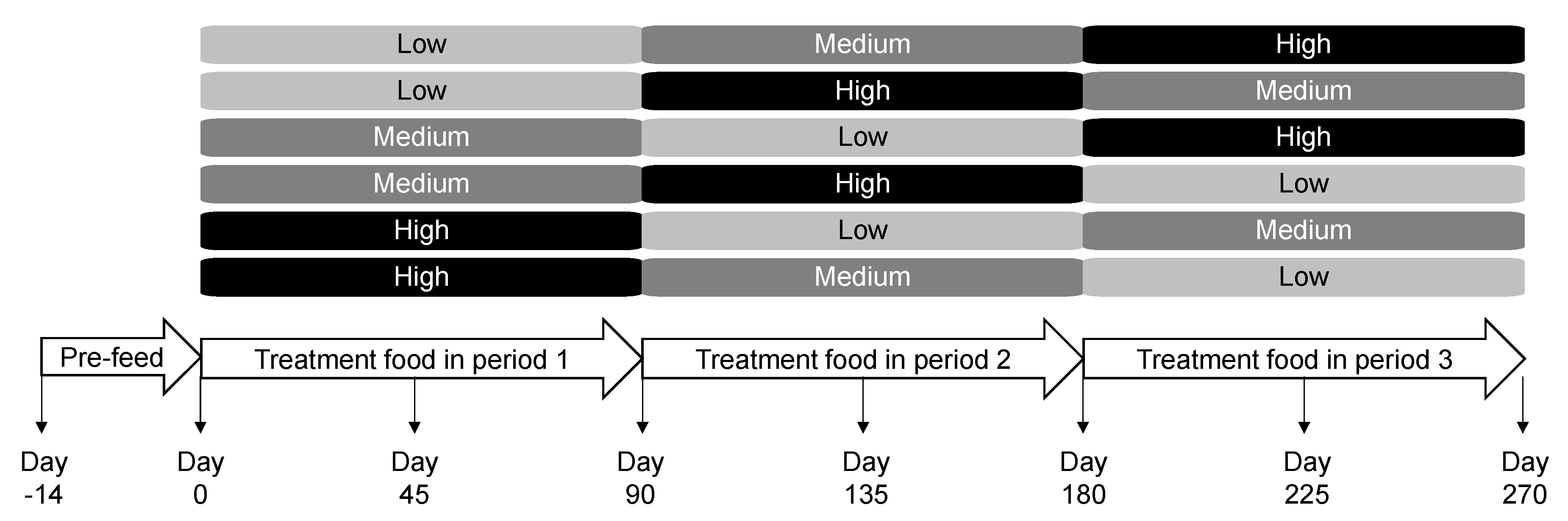
4.2. Animals and Experimental Design
4.3. Stool Scoring and Fecal Sample Processing
4.4. Metabolite Analyses
4.5. Fecal Microbiome Analysis and Bioinformatics Processing
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Association of American Feed Control Officials. AAFCO Methods for Substantiating Nutritional Adequacy of Dog and Cat Foods. Available online: https://www.aafco.org/Portals/0/SiteContent/Regulatory/Committees/Pet-Food/Reports/Pet_Food_Report_2013_Midyear-Proposed_Revisions_to_AAFCO_Nutrient_Profiles.pdf (accessed on 7 October 2019).
- Hang, I.; Rinttila, T.; Zentek, J.; Kettunen, A.; Alaja, S.; Apajalahti, J.; Harmoinen, J.; de Vos, W.M.; Spillmann, T. Effect of high contents of dietary animal-derived protein or carbohydrates on canine faecal microbiota. BMC Vet. Res. 2012, 8, 90. [Google Scholar] [CrossRef] [Green Version]
- Bermingham, E.N.; Maclean, P.; Thomas, D.G.; Cave, N.J.; Young, W. Key bacterial families (Clostridiaceae, Erysipelotrichaceae and Bacteroidaceae) are related to the digestion of protein and energy in dogs. PeerJ 2017, 5, e3019. [Google Scholar] [CrossRef]
- Hang, I.; Heilmann, R.M.; Grutzner, N.; Suchodolski, J.S.; Steiner, J.M.; Atroshi, F.; Sankari, S.; Kettunen, A.; de Vos, W.M.; Zentek, J.; et al. Impact of diets with a high content of greaves-meal protein or carbohydrates on faecal characteristics, volatile fatty acids and faecal calprotectin concentrations in healthy dogs. BMC Vet. Res. 2013, 9, 201. [Google Scholar] [CrossRef] [Green Version]
- Herstad, K.M.V.; Gajardo, K.; Bakke, A.M.; Moe, L.; Ludvigsen, J.; Rudi, K.; Rud, I.; Sekelja, M.; Skancke, E. A diet change from dry food to beef induces reversible changes on the faecal microbiota in healthy, adult client-owned dogs. BMC Vet. Res. 2017, 13, 147. [Google Scholar] [CrossRef]
- Andre, A.; Leriche, I.; Chaix, G.; Thorin, C.; Burger, M.; Nguyen, P. Recovery of insulin sensitivity and optimal body composition after rapid weight loss in obese dogs fed a high-protein medium-carbohydrate diet. J. Anim. Physiol. Anim. Nutr. 2017, 101, 21–30. [Google Scholar] [CrossRef] [Green Version]
- Coelho, L.P.; Kultima, J.R.; Costea, P.I.; Fournier, C.; Pan, Y.; Czarnecki-Maulden, G.; Hayward, M.R.; Forslund, S.K.; Schmidt, T.S.B.; Descombes, P.; et al. Similarity of the dog and human gut microbiomes in gene content and response to diet. Microbiome 2018, 6, 72. [Google Scholar] [CrossRef]
- Li, Q.; Lauber, C.L.; Czarnecki-Maulden, G.; Pan, Y.; Hannah, S.S. Effects of the dietary protein and carbohydrate ratio on gut microbiomes in dogs of different body conditions. MBio 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Verbrugghe, A.; Lourenco, M.; Cools, A.; Liu, D.J.X.; van de Wiele, T.; Marzorati, M.; Eeckhaut, V.; van Immerseel, F.; Vanhaecke, L.; et al. The response of canine faecal microbiota to increased dietary protein is influenced by body condition. BMC Vet. Res. 2017, 13, 374. [Google Scholar] [CrossRef] [Green Version]
- Jackson, M.I.; Jewell, D.E. Balance of saccharolysis and proteolysis underpins improvements in stool quality induced by adding a fiber bundle containing bound polyphenols to either hydrolyzed meat or grain-rich foods. Gut Microbes 2019, 10, 298–320. [Google Scholar] [CrossRef] [Green Version]
- Pinna, C.; Vecchiato, C.G.; Bolduan, C.; Grandi, M.; Stefanelli, C.; Windisch, W.; Zaghini, G.; Biagi, G. Influence of dietary protein and fructooligosaccharides on fecal fermentative end-products, fecal bacterial populations and apparent total tract digestibility in dogs. BMC Vet. Res. 2018, 14, 106. [Google Scholar] [CrossRef] [Green Version]
- Nery, J.; Goudez, R.; Biourge, V.; Tournier, C.; Leray, V.; Martin, L.; Thorin, C.; Nguyen, P.; Dumon, H. Influence of dietary protein content and source on colonic fermentative activity in dogs differing in body size and digestive tolerance. J. Anim. Sci. 2012, 90, 2570–2580. [Google Scholar] [CrossRef]
- Martinez, K.B.; Leone, V.; Chang, E.B. Microbial metabolites in health and disease: Navigating the unknown in search of function. J. Biol. Chem. 2017, 292, 8553–8559. [Google Scholar] [CrossRef] [Green Version]
- Heintz-Buschart, A.; Wilmes, P. Human gut microbiome: Function matters. Trends Microbiol. 2018, 26, 563–574. [Google Scholar] [CrossRef]
- Matsumoto, M.; Ooga, T.; Kibe, R.; Aiba, Y.; Koga, Y.; Benno, Y. Colonic absorption of low-molecular-weight metabolites influenced by the intestinal microbiome: A pilot study. PLoS ONE 2017, 12, e0169207. [Google Scholar] [CrossRef]
- Ephraim, E.; Jackson, M.I.; Yerramilli, M.; Jewell, D.E. Soluble fiber and omega-3 fatty acids reduce levels of advanced glycation end products and uremic toxins in senior dogs by modulating the gut microbiome. J. Food Sci. Nutr. Res. 2020, 3, 18–33. [Google Scholar] [CrossRef]
- Weiner, I.D.; Mitch, W.E.; Sands, J.M. Urea and ammonia metabolism and the control of renal nitrogen excretion. Clin. J. Am. Soc. Nephrol. 2015, 10, 1444–1458. [Google Scholar] [CrossRef] [Green Version]
- Aronov, P.A.; Luo, F.J.; Plummer, N.S.; Quan, Z.; Holmes, S.; Hostetter, T.H.; Meyer, T.W. Colonic contribution to uremic solutes. J. Am. Soc. Nephrol. 2011, 22, 1769–1776. [Google Scholar] [CrossRef] [Green Version]
- Davies, R. The metabolomic quest for a biomarker in chronic kidney disease. Clin. Kidney J. 2018, 11, 694–703. [Google Scholar] [CrossRef]
- Sun, C.Y.; Hsu, H.H.; Wu, M.S. p-Cresol sulfate and indoxyl sulfate induce similar cellular inflammatory gene expressions in cultured proximal renal tubular cells. Nephrol. Dial. Transpl. 2013, 28, 70–78. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.; He, F.; Wu, C.; Li, P.; Li, N.; Deng, J.; Zhu, G.; Ren, W.; Peng, Y. Betaine in inflammation: Mechanistic aspects and applications. Front. Immunol. 2018, 9, 1070. [Google Scholar] [CrossRef] [Green Version]
- Koistinen, V.M.; Kärkkäinen, O.; Borewicz, K.; Zarei, I.; Jokkala, J.; Micard, V.; Rosa-Sibakov, N.; Auriola, S.; Aura, A.M.; Smidt, H.; et al. Contribution of gut microbiota to metabolism of dietary glycine betaine in mice and in vitro colonic fermentation. Microbiome 2019, 7, 103. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Wang, Z.; Greenwald, J.; Kothapalli, K.S.; Park, H.G.; Liu, R.; Mendralla, E.; Lawrence, P.; Wang, X.; Brenna, J.T. BCFA suppresses LPS induced IL-8 mRNA expression in human intestinal epithelial cells. Prostaglandins Leukot. Essent. Fat. Acids 2017, 116, 27–31. [Google Scholar] [CrossRef]
- Wongtangtintharn, S.; Oku, H.; Iwasaki, H.; Toda, T. Effect of branched-chain fatty acids on fatty acid biosynthesis of human breast cancer cells. J. Nutr. Sci. Vitam. 2004, 50, 137–143. [Google Scholar] [CrossRef] [Green Version]
- Ran-Ressler, R.R.; Khailova, L.; Arganbright, K.M.; Adkins-Rieck, C.K.; Jouni, Z.E.; Koren, O.; Ley, R.E.; Brenna, J.T.; Dvorak, B. Branched chain fatty acids reduce the incidence of necrotizing enterocolitis and alter gastrointestinal microbial ecology in a neonatal rat model. PLoS ONE 2011, 6, e29032. [Google Scholar] [CrossRef] [Green Version]
- Macfarlane, G.T.; Gibson, G.R.; Beatty, E.; Cummings, J.H. Estimation of short-chain fatty acid production from protein by human intestinal bacteria based on branched-chain fatty acid measurements. FEMS Microbiol. Ecol. 1992, 101, 81–88. [Google Scholar]
- Abdul Rahim, M.B.H.; Chilloux, J.; Martinez-Gili, L.; Neves, A.L.; Myridakis, A.; Gooderham, N.; Dumas, M.E. Diet-induced metabolic changes of the human gut microbiome: Importance of short-chain fatty acids, methylamines and indoles. Acta Diabetol. 2019, 56, 493–500. [Google Scholar] [CrossRef] [Green Version]
- Levan, S.R.; Stamnes, K.A.; Lin, D.L.; Panzer, A.R.; Fukui, E.; McCauley, K.; Fujimura, K.E.; McKean, M.; Ownby, D.R.; Zoratti, E.M.; et al. Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat. Microbiol. 2019, 4, 1851–1861. [Google Scholar] [CrossRef]
- Fehlner-Peach, H.; Magnabosco, C.; Raghavan, V.; Scher, J.U.; Tett, A.; Cox, L.M.; Gottsegen, C.; Watters, A.; Wiltshire-Gordon, J.D.; Segata, N.; et al. Distinct polysaccharide utilization profiles of human intestinal Prevotella copri isolates. Cell Host Microbe 2019, 26, 680–690.e685. [Google Scholar] [CrossRef]
- Crost, E.H.; le Gall, G.; Laverde-Gomez, J.A.; Mukhopadhya, I.; Flint, H.J.; Juge, N. Mechanistic insights into the cross-feeding of Ruminococcus gnavus and Ruminococcus bromii on host and dietary carbohydrates. Front. Microbiol. 2018, 9, 2558. [Google Scholar] [CrossRef] [Green Version]
- Kaakoush, N.O. Insights into the role of Erysipelotrichaceae in the human host. Front. Cell Infect. Microbiol. 2015, 5, 84. [Google Scholar] [CrossRef] [Green Version]
- Hugenholtz, F.; Davids, M.; Schwarz, J.; Muller, M.; Tome, D.; Schaap, P.; Hooiveld, G.; Smidt, H.; Kleerebezem, M. Metatranscriptome analysis of the microbial fermentation of dietary milk proteins in the murine gut. PLoS ONE 2018, 13, e0194066. [Google Scholar] [CrossRef] [Green Version]
- Martin, R.; Miquel, S.; Benevides, L.; Bridonneau, C.; Robert, V.; Hudault, S.; Chain, F.; Berteau, O.; Azevedo, V.; Chatel, J.M.; et al. Functional characterization of novel Faecalibacterium prausnitzii strains isolated from healthy volunteers: A step forward in the use of F. prausnitzii as a next-generation probiotic. Front. Microbiol. 2017, 8, 1226. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [Green Version]
- Rajilic-Stojanovic, M.; Biagi, E.; Heilig, H.G.; Kajander, K.; Kekkonen, R.A.; Tims, S.; de Vos, W.M. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 2011, 141, 1792–1801. [Google Scholar] [CrossRef]
- Suchodolski, J.S.; Markel, M.E.; Garcia-Mazcorro, J.F.; Unterer, S.; Heilmann, R.M.; Dowd, S.E.; Kachroo, P.; Ivanov, I.; Minamoto, Y.; Dillman, E.M.; et al. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS ONE 2012, 7, e51907. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016, 535, 376–381. [Google Scholar] [CrossRef]
- Pianta, A.; Arvikar, S.; Strle, K.; Drouin, E.E.; Wang, Q.; Costello, C.E.; Steere, A.C. Evidence of the immune relevance of Prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol. 2017, 69, 964–975. [Google Scholar] [CrossRef] [Green Version]
- De Filippis, F.; Pasolli, E.; Tett, A.; Tarallo, S.; Naccarati, A.; de Angelis, M.; Neviani, E.; Cocolin, L.; Gobbetti, M.; Segata, N.; et al. Distinct genetic and functional traits of human intestinal Prevotella copri strains are associated with different habitual diets. Cell Host Microbe 2019, 25, 444–453.e443. [Google Scholar] [CrossRef] [Green Version]
- Balty, C.; Guillot, A.; Fradale, L.; Brewee, C.; Boulay, M.; Kubiak, X.; Benjdia, A.; Berteau, O. Ruminococcin C, an anti-clostridial sactipeptide produced by a prominent member of the human microbiota Ruminococcus Gnavus. J. Biol. Chem. 2019, 294, 14512–14525. [Google Scholar] [CrossRef] [Green Version]
- Henke, M.T.; Kenny, D.J.; Cassilly, C.D.; Vlamakis, H.; Xavier, R.J.; Clardy, J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. USA 2019, 116, 12672–12677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeffery, I.B.; Das, A.; O’Herlihy, E.; Coughlan, S.; Cisek, K.; Moore, M.; Bradley, F.; Carty, T.; Pradhan, M.; Dwibedi, C.; et al. Differences in fecal microbiomes and metabolomes of people with vs without irritable bowel syndrome and bile acid malabsorption. Gastroenterology 2020, 158, 1016–1028.e8. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.A.; Jewell, D.E. Feeding healthy beagles medium-chain triglycerides, fish oil, and carnitine offsets age-related changes in serum fatty acids and carnitine metabolites. PLoS ONE 2012, 7, e49510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, J.A.; Jackson, M.I.; Farace, G.; Yerramilli, M.; Jewell, D.E. Influence of dietary ingredients on lean body percent, uremic toxin concentrations, and kidney function in senior-adult cats. Metabolites 2019, 9. [Google Scholar] [CrossRef] [Green Version]
- Golder, C.; Weemhoff, J.L.; Jewell, D.E. Cats have increased protein digestibility as compared to dogs and improve their ability to absorb protein as dietary protein intake shifts from animal to plant sources. Animals 2020, 10, 541. [Google Scholar] [CrossRef] [Green Version]
- National Research Council Committee. Update of the Guide for the Care and Use of Laboratory Animals, 8th ed.; The National Academies Collection: Reports funded by National Institutes of Health; National Academies Press: Washington, DC, USA, 2011. [Google Scholar] [CrossRef]
- Wang, B.-S.; Wang, X.-J.; Gong, L.-K. The construction of a Williams Design and randomization in cross-over clinical trials using SAS. J. Stat. Softw. 2009, 29, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Hall, J.A.; Melendez, L.D.; Jewell, D.E. Using gross energy improves metabolizable energy predictive equations for pet foods whereas undigested protein and fiber content predict stool quality. PLoS ONE 2013, 8, e54405. [Google Scholar] [CrossRef] [Green Version]
- Hall, J.A.; Yerramilli, M.; Obare, E.; Yerramilli, M.; Yu, S.; Jewell, D.E. Comparison of serum concentrations of symmetric dimethylarginine and creatinine as kidney function biomarkers in healthy geriatric cats fed reduced protein foods enriched with fish oil, L-carnitine, and medium-chain triglycerides. Vet. J. 2014, 202, 588–596. [Google Scholar] [CrossRef]
- Yi, N.; Tang, Z.; Zhang, X.; Guo, B. BhGLM: Bayesian hierarchical GLMs and survival models, with applications to genomics and epidemiology. Bioinformatics 2019, 35, 1419–1421. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.4-6. Available online: https://CRAN.R-project.org/package=vegan (accessed on 20 January 2019).
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ephraim, E.; Cochrane, C.-Y.; Jewell, D.E. Varying Protein Levels Influence Metabolomics and the Gut Microbiome in Healthy Adult Dogs. Toxins 2020, 12, 517. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12080517
Ephraim E, Cochrane C-Y, Jewell DE. Varying Protein Levels Influence Metabolomics and the Gut Microbiome in Healthy Adult Dogs. Toxins. 2020; 12(8):517. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12080517
Chicago/Turabian StyleEphraim, Eden, Chun-Yen Cochrane, and Dennis E. Jewell. 2020. "Varying Protein Levels Influence Metabolomics and the Gut Microbiome in Healthy Adult Dogs" Toxins 12, no. 8: 517. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12080517




