Development of Anti-Idiotypic Nanobody-Phage Based Immuno-Loop-Mediated Isothermal Amplification Assay for Aflatoxins in Peanuts
Abstract
:1. Introduction
2. Results
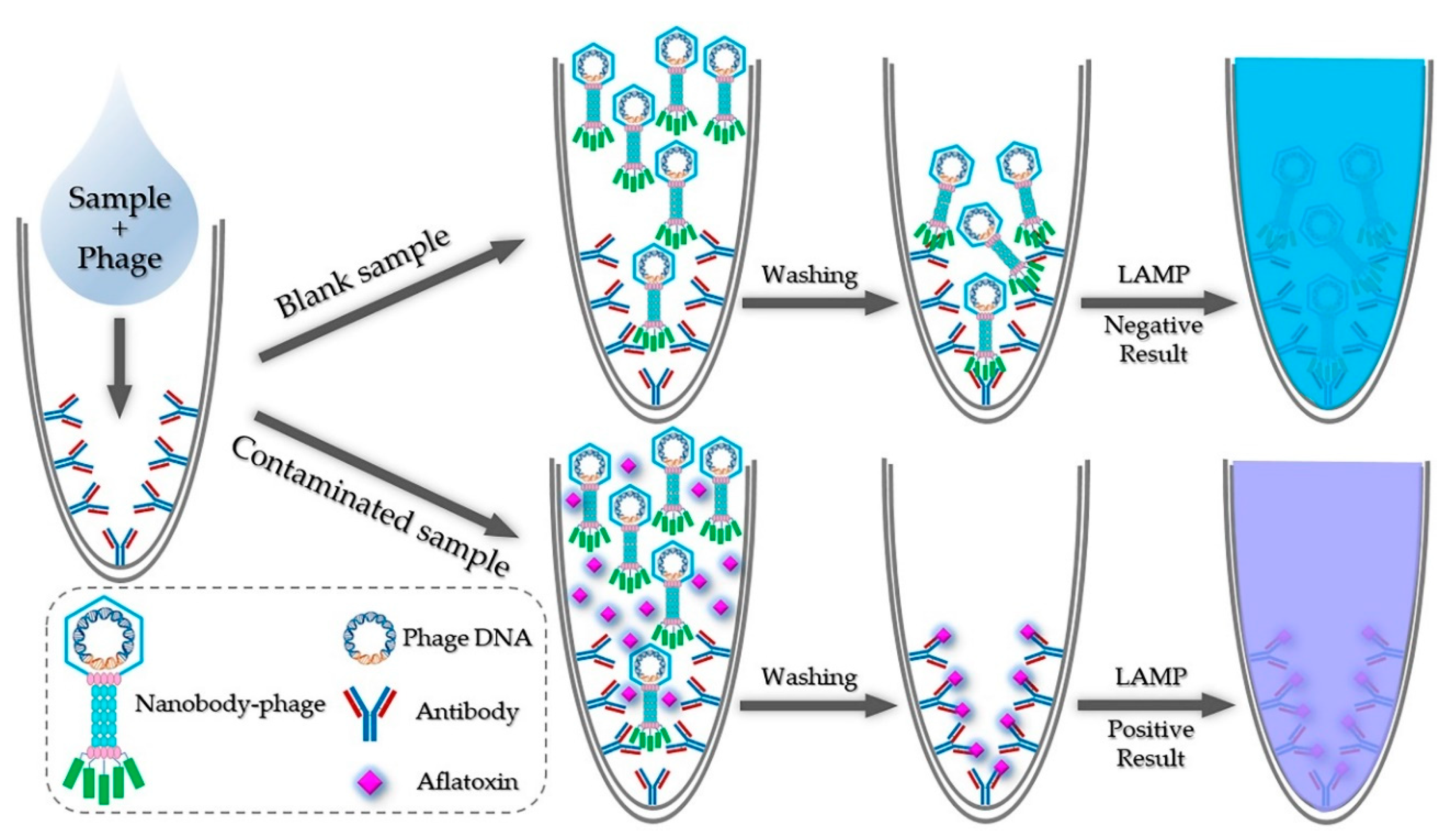
2.1. Principle of iLAMP
2.2. LAMP Primers
2.3. Specificity of LAMP
2.4. Visual Inspection of LAMP
2.5. Visual Detection Limit of iLAMP
2.6. Solvent and Matrix Effects
2.7. Assay Validation with Peanut Samples
2.8. Comparison of iLAMP with Previous Research
3. Discussion
3.1. Primer Design of LAMP
3.2. Visual Inspection Indicator of LAMP
3.3. Solvent and Matrix Affects
3.4. Application Prospect of iLAMP
4. Materials and Methods
4.1. Materials
4.2. Phage Preparation
4.3. Primer Design
4.4. LAMP Reaction
4.5. iLAMP Method
4.6. Sample Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Yabe, K.; Ando, Y.; Hamasaki, T. Biosynthetic relationship among aflatoxins B1, B2, G1, and G2. Appl. Environ. Microbiol. 1988, 54, 2101–2106. [Google Scholar] [CrossRef] [Green Version]
- Some Traditional Herbal Medicines, Some Mycotoxins, Naphthalene and Styrene; World Health Organization: Geneva, Switzerland, 2002.
- Kensler, T.W.; Roebuck, B.D.; Wogan, G.N.; Groopman, J.D. Aflatoxin: A 50-year odyssey of mechanistic and translational toxicology. Toxicol. Sci. 2011, 120, S28–S48. [Google Scholar] [CrossRef] [Green Version]
- Ismail, A.; Goncalves, B.L.; de Neeff, D.V.; Ponzilacqua, B.; Coppa, C.; Hintzsche, H.; Sajid, M.; Cruz, A.G.; Corassin, C.H.; Oliveira, C.A.F. Aflatoxin in foodstuffs: Occurrence and recent advances in decontamination. Food Res. Int. 2018, 113, 74–85. [Google Scholar] [CrossRef]
- Yakubu, A.; Vyas, A. Aflatoxin: Occurrence, Regulation, and Detection in Food and Feed. In Microbial Biotechnology: Basic Research and Applications; Singh, J., Vyas, A., Wang, S., Prasad, R., Eds.; Springer: Singapore, 2020; pp. 337–353. [Google Scholar] [CrossRef]
- Shephard, G.S. Aflatoxin analysis at the beginning of the twenty-first century. Anal. Bioanal. Chem. 2009, 395, 1215–1224. [Google Scholar] [CrossRef]
- Li, P.; Zhang, Q.; Zhang, W.; Zhang, J.; Chen, X.; Jiang, J.; Xie, L.; Zhang, D. Development of a class-specific monoclonal antibody-based ELISA for aflatoxins in peanut. Food Chem. 2009, 115, 313–317. [Google Scholar] [CrossRef]
- Zhang, D.; Li, P.; Yang, Y.; Zhang, Q.; Zhang, W.; Xiao, Z.; Ding, X. A high selective immunochromatographic assay for rapid detection of aflatoxin B 1. Talanta 2011, 85, 736–742. [Google Scholar] [CrossRef]
- Ma, H.; Sun, J.; Zhang, Y.; Bian, C.; Xia, S.; Zhen, T. Label-free immunosensor based on one-step electrodeposition of chitosan-gold nanoparticles biocompatible film on Au microelectrode for determination of aflatoxin B1 in maize. Biosens. Bioelectron. 2016, 80, 222–229. [Google Scholar] [CrossRef]
- Berg, E.A.; Fishman, J.B. Labeling Antibodies. Cold Spring Harb. Protoc. 2020, 2020, 099242. [Google Scholar] [CrossRef]
- Li, P.; Zhang, Q.; Zhang, W. Immunoassays for aflatoxins. TrAC Trends Anal. Chem. 2009, 28, 1115–1126. [Google Scholar] [CrossRef]
- Wang, Y.; Li, P.; Majkova, Z.; Bever, C.R.; Kim, H.J.; Zhang, Q.; Dechant, J.E.; Gee, S.J.; Hammock, B.D. Isolation of Alpaca Anti-Idiotypic Heavy-Chain Single-Domain Antibody for the Aflatoxin Immunoassay. Anal. Chem. 2013, 85, 8298–8303. [Google Scholar] [CrossRef] [Green Version]
- Lei, J.; Li, P.; Zhang, Q.; Wang, Y.; Zhang, Z.; Ding, X.; Zang, W. Anti-Idiotypic Nanobody-Phage Based Real-Time Immuno-PCR for Detection of Hepatocarcinogen Aflatoxin in Grains and Feedstuffs. Anal. Chem. 2014, 86, 10841–10846. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [Green Version]
- Sagcan, H.; Kara, N.T. Detection of Potato ring rot Pathogen Clavibacter michiganensis subsp. s epedonicus by Loop-mediated isothermal amplification (LAMP) assay. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Rivero, R.; Bisio, M.; Velázquez, E.B.; Esteva, M.I.; Scollo, K.; González, N.L.; Altcheh, J.; Ruiz, A.M. Rapid detection of Trypanosoma cruzi by colorimetric loop-mediated isothermal amplification (LAMP): A potential novel tool for the detection of congenital Chagas infection. Diagn. Microbiol. Infect. Dis. 2017, 89, 26–28. [Google Scholar] [CrossRef]
- Li, R.; Shi, J.; Liu, B.; Zhang, D.; Zhao, X.; Yang, L. International collaborative ring trial of four gene-specific loop-mediated isothermal amplification assays in GMO analysis. Food Control 2018, 84, 278–283. [Google Scholar] [CrossRef]
- Niessen, L.; Bechtner, J.; Fodil, S.; Taniwaki, M.H.; Vogel, R.F. LAMP-based group specific detection of aflatoxin producers within Aspergillus section Flavi in food raw materials, spices, and dried fruit using neutral red for visible-light signal detection. Int. J. Food Microbiol. 2018, 266, 241–250. [Google Scholar] [CrossRef]
- Kundapur, R.R.; Nema, V. Loop-mediated isothermal amplification: Beyond microbial identification. Cogent Biol. 2016, 2, 1137110. [Google Scholar] [CrossRef]
- Nurul Najian, A.B.; Foo, P.C.; Ismail, N.; Kim-Fatt, L.; Yean, C.Y. Probe-specific loop-mediated isothermal amplification magnetogenosensor assay for rapid and specific detection of pathogenic Leptospira. Mol. Cell. Probes 2019, 44, 63–68. [Google Scholar] [CrossRef]
- Zhu, L.; Xu, Y.; Cheng, N.; Xie, P.; Shao, X.; Huang, K.; Luo, Y.; Xu, W. A facile cascade signal amplification strategy using DNAzyme loop-mediated isothermal amplification for the ultrasensitive colorimetric detection of Salmonella. Sens. Actuators B Chem. 2017, 242, 880–888. [Google Scholar] [CrossRef]
- Reid, M.S.; Le, X.C.; Zhang, H. Exponential Isothermal Amplification of Nucleic Acids and Assays for Proteins, Cells, Small Molecules, and Enzyme Activities: An EXPAR Example. Angew. Chem. Int. Ed. 2018, 57, 11856–11866. [Google Scholar] [CrossRef]
- Pourhassan-Moghaddam, M.; Rahmati-Yamchi, M.; Akbarzadeh, A.; Daraee, H.; Nejati-Koshki, K.; Hanifehpour, Y.; Joo, S. Protein detection through different platforms of immuno-loop-mediated isothermal amplification. Nanoscale Res. Lett. 2013, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Iwamoto, T.; Sonobe, T.; Hayashi, K. Loop-Mediated Isothermal Amplification for Direct Detection of Mycobacterium tuberculosis Complex, M. avium, and M. intracellulare in Sputum Samples. J. Clin. Microbiol. 2003, 41, 2616–2622. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.H.; Li, P.W.; Zhang, Q.; Zhang, W.; Huang, Y.L.; Ding, X.X.; Jiang, J. Production of ultrasensitive generic monoclonal antibodies against major aflatoxins using a modified two-step screening procedure. Anal. Chim. Acta. 2009, 636, 63–69. [Google Scholar] [CrossRef]
- Li, X.; Li, P.; Lei, J.; Zhang, Q.; Zhang, W.; Li, C. A simple strategy to obtain ultra-sensitive single-chain fragment variable antibodies for aflatoxin detection. RSC Adv. 2013, 3, 22367–22372. [Google Scholar] [CrossRef]
- Zhang, D.H.; Li, P.W.; Zhang, Q.; Zhang, W. Ultrasensitive nanogold probe-based immunochromatographic assay for simultaneous detection of total aflatoxins in peanuts. Biosens. Bioelectron. 2011, 26, 2877–2882. [Google Scholar] [CrossRef]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef]
- Apollonio, L.G.; Whittall, I.R.; Pianca, D.J.; Kyd, J.M.; Maher, W.A. Matrix effect and cross-reactivity of select amphetamine-type substances, designer analogues, and putrefactive amines using the Bio-Quant direct ELISA presumptive assays for amphetamine and methamphetamine. J. Anal. Toxicol. 2007, 31, 208–213. [Google Scholar] [CrossRef]
- Vergara, A.; Boutal, H.; Ceccato, A.; López, M.; Cruells, A.; Bueno-Freire, L.; Moreno-Morales, J.; Puig de la Bellacasa, J.; Castro, P.; Torres, A. Assessment of a loop-mediated isothermal amplification (LAMP) assay for the rapid detection of pathogenic bacteria from respiratory samples in patients with hospital-acquired pneumonia. Microorganisms 2020, 8, 103. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-G.; Baek, S.H.; Kim, S.; Kim, H.I.; Lee, S.W.; Kailasa, S.K.; Park, T.J. Rapid discriminative detection of dengue viruses via loop mediated isothermal amplification. Talanta 2018, 190, 391–396. [Google Scholar] [CrossRef]
- Kanitkar, Y.H.; Stedtfeld, R.D.; Steffan, R.J.; Hashsham, S.A.; Cupples, A.M. Loop-mediated isothermal amplification (LAMP) for rapid detection and quantification of Dehalococcoides biomarker genes in commercial reductive dechlorinating cultures KB-1 and SDC-9. Appl. Environ. Microbiol. 2016, 82, 1799–1806. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Xu, Y.; Xiong, Y.H.; Tu, Z.; Li, Y.P.; He, Z.Y.; Qiu, Y.L.; Fu, J.H.; Gee, S.J.; Hammock, B.D. VHH phage-based competitive real-time immuno-polymerase chain reaction for ultrasensitive detection of ochratoxin A in cereal. Anal. Chem. 2014, 86, 7471–7477. [Google Scholar] [CrossRef]
- Ma, F.; Chen, R.; Li, P.; Zhang, Q.; Zhang, W.; Hu, X. Preparation of an immunoaffinity column with amino-silica gel microparticles and its application in sample cleanup for aflatoxin detection in agri-products. Molecules 2013, 18, 2222–2235. [Google Scholar] [CrossRef]






| Sample | HPLC (µg/kg) | iLAMP (n = 5) | |||
|---|---|---|---|---|---|
| AFB1 | AFB2 | AFG1 | AFG2 | ||
| 1 | 3.39 | 0.20 | ND a | ND | V b, V, V, V, V |
| 2 | 3.73 | 0.36 | ND | ND | V, V, V, V, V |
| 3 | 2.27 | 0.22 | ND | ND | V, V, V, V, V |
| 4 | 23.60 | 3.60 | ND | ND | V, V, V, V, V |
| 5 | 2.56 | 0.31 | ND | ND | V, V, V, V, V |
| 6 | 12.41 | 2.30 | ND | ND | V, V, V, V, V |
| 7 | 0.78 | ND | ND | ND | S c, S, S, S, S |
| 8 | 4.90 | 0.61 | ND | ND | V, V, V, V, V |
| 9 | 1.97 | ND | ND | ND | V, V, V, V, V |
| 10 | 20.68 | 1.66 | ND | ND | V, V, V, V, V |
| 11 | 1.02 | ND | ND | ND | S, S, S, S, S |
| 12 | 6.24 | 1.02 | ND | ND | V, V, V, V, V |
| 13 | 16.43 | 1.92 | 2.36 | ND | V, V, V, V, V |
| 14 | 0.94 | ND | ND | ND | S, S, S, S, S |
| 15 | 16.84 | 3.32 | ND | ND | V, V, V, V, V |
| 16 | 28.86 | 1.30 | ND | ND | V, V, V, V, V |
| 17 | 2.24 | 0.44 | ND | ND | V, V, V, V, V |
| 18 | 15.88 | 2.25 | ND | ND | V, V, V, V, V |
| 19 | 5.18 | 0.66 | ND | ND | V, V, V, V, V |
| 20 | 0.34 | ND | ND | ND | S, S, S, S, S |
| Method | Immune Reagents | Matrix | Sensitivity (µg/kg) | Reference |
|---|---|---|---|---|
| ELISA | 1C11 + AFB1-BSA | Standard solution | 0.0012 | [25] |
| ICA | 1C11 + AFB1-OVA | Peanut | 0.03 | [27] |
| ELISA | 1C11 + Nanobody | Peanut, corn, rice | 13.8 | [12] |
| iPCR | 1C11 + Nanobody-phage | Peanut, corn, rice, feedstuff | 5.6 | [13] |
| iLAMP | 1C11 + Nanobody-phage | Peanut | 1.6 | _ a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, J.; Han, X.; Tang, X.; Wang, H.; Zhang, Q. Development of Anti-Idiotypic Nanobody-Phage Based Immuno-Loop-Mediated Isothermal Amplification Assay for Aflatoxins in Peanuts. Toxins 2020, 12, 565. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12090565
Lei J, Han X, Tang X, Wang H, Zhang Q. Development of Anti-Idiotypic Nanobody-Phage Based Immuno-Loop-Mediated Isothermal Amplification Assay for Aflatoxins in Peanuts. Toxins. 2020; 12(9):565. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12090565
Chicago/Turabian StyleLei, Jiawen, Xiaole Han, Xiaoqian Tang, Haiying Wang, and Qi Zhang. 2020. "Development of Anti-Idiotypic Nanobody-Phage Based Immuno-Loop-Mediated Isothermal Amplification Assay for Aflatoxins in Peanuts" Toxins 12, no. 9: 565. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12090565





