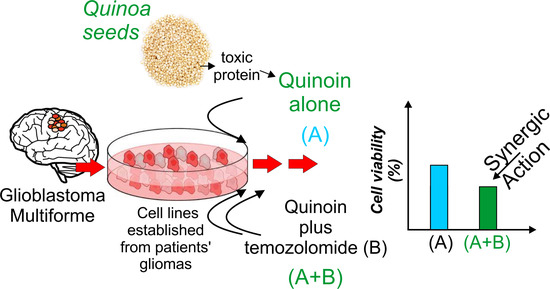
Cytotoxicity Effect of Quinoin, Type 1 Ribosome-Inactivating Protein from Quinoa Seeds, on Glioblastoma Cells
Abstract
:1. Introduction
2. Results
2.1. Quinoin Isolation
2.2. Inhibiting Effect of Quinoin on Cell Growth and Viability of Glioblastoma U87Mg and Patient-Derived Cell Lines NULU and ZAR
2.3. Quinoin Treatment Results in Morphological Alteration in U87Mg Cells
2.4. Quinoin and Oxidative Stress
2.5. Quinoin as a Potential Adjuvant for Glioblastoma Treatment in Combination with Temozolomide
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Quinoin Purification
4.3. Cell Culture
4.4. IC50 Estimation of Quinoin and Temozolomide in U87Mg and Patient-Derived GBM Cell Lines
4.5. Proliferation Assay
4.6. Cell Viability Test
4.7. Microscopic Observation of Live Cells
4.8. Combined Treatment with TMZ and Quinoin
4.9. Western Blot Analysis of U87Mg Cells Treated with Quinoin
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stirpe, F. Ribosome-inactivating proteins: From toxins to useful proteins. Toxicon 2013, 67, 12–16. [Google Scholar] [CrossRef]
- Endo, Y.; Tsurugi, K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J. Biol. Chem. 1987, 262, 8128–8130. [Google Scholar] [CrossRef]
- Shi, X.; Khade, P.K.; Sanbonmatsu, K.Y.; Joseph, S. Functional role of the sarcin-ricin loop of the 23S rRNA in the elongation cycle of protein synthesis. J. Mol. Biol. 2012, 419, 125–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbieri, L.; Battelli, M.G.; Stirpe, F. Ribosome-inactivating proteins from plants. Biochim. Biophys. Acta 1993, 1154, 237–282. [Google Scholar] [CrossRef]
- Tejero, J.; Jiménez, P.; Quinto, E.J.; Cordoba-Diaz, D.; Garrosa, M.; Cordoba-Diaz, M.; Gayoso, M.J.; Girbés, T. Elderberries: A source of ribosome-inactivating proteins with lectin activity. Molecules 2015, 20, 2364–2387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Zaeytijd, J.; Van Damme, E.J. Extensive Evolution of Cereal Ribosome-Inactivating Proteins Translates into Unique Structural Features, Activation Mechanisms, and Physiological Roles. Toxins 2017, 9, 123. [Google Scholar] [CrossRef]
- Park, S.W.; Lawrence, C.B.; Linden, J.C.; Vivanco, J.M. Isolation and characterization of a novel ribosome-inactivating protein from root cultures of pokeweed and its mechanism of secretion from roots. Plant Physiol. 2002, 130, 164–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Maro, A.; Citores, L.; Russo, R.; Iglesias, R.; Ferreras, J.M. Sequence comparison and phylogenetic analysis by the Maximum Likelihood method of ribosome-inactivating proteins from angiosperms. Plant Mol. Biol. 2014, 85, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Lapadula, W.J.; Ayub, M.J. Ribosome Inactivating Proteins from an evolutionary perspective. Toxicon 2017, 136, 6–14. [Google Scholar] [CrossRef]
- Zhu, F.; Zhou, Y.K.; Ji, Z.L.; Chen, X.R. The Plant Ribosome-Inactivating Proteins Play Important Roles in Defense against Pathogens and Insect Pest Attacks. Front. Plant Sci. 2018, 9, 146. [Google Scholar] [CrossRef] [Green Version]
- Aceto, S.; Di Maro, A.; Conforto, B.; Siniscalco, G.G.; Parente, A.; Delli Bovi, P.; Gaudio, L. Nicking activity on pBR322 DNA of ribosome inactivating proteins from Phytolacca dioica L. leaves. Biol. Chem. 2005, 386, 307–317. [Google Scholar] [CrossRef]
- Pizzo, E.; Di Maro, A. A new age for biomedical applications of Ribosome Inactivating Proteins (RIPs): From bioconjugate to nanoconstructs. J. Biomed. Sci. 2016, 23, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Virgilio, M.; Lombardi, A.; Caliandro, R.; Fabbrini, M.S. Ribosome-inactivating proteins: From plant defense to tumor attack. Toxins 2010, 2, 2699–2737. [Google Scholar] [CrossRef] [Green Version]
- Rust, A.; Partridge, L.J.; Davletov, B.; Hautbergue, G.M. The Use of Plant-Derived Ribosome Inactivating Proteins in Immunotoxin Development: Past, Present and Future Generations. Toxins 2017, 9, 334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stirpe, F.; Gilabert-Oriol, R. Ribosome-Inactivating Proteins: An Overview. In Plant Toxins; Carlini, C.R., Ligabue-Braun, R., Gopalakrishnakone, P., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 153–182. [Google Scholar]
- Akkouh, O.; Ng, T.B.; Cheung, R.C.; Wong, J.H.; Pan, W.; Ng, C.C.; Sha, O.; Shaw, P.C.; Chan, W.Y. Biological activities of ribosome-inactivating proteins and their possible applications as antimicrobial, anticancer, and anti-pest agents and in neuroscience research. Appl. Microbiol. Biotechnol. 2015, 99, 9847–9863. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.Q.; Zhu, Z.N.; Zheng, Y.T.; Shaw, P.C. Engineering of Ribosome-inactivating Proteins for Improving Pharmacological Properties. Toxins 2020, 12, 167. [Google Scholar] [CrossRef] [Green Version]
- Miao, J.; Jiang, Y.; Wang, D.; Zhou, J.; Fan, C.; Jiao, F.; Liu, B.; Zhang, J.; Wang, Y.; Zhang, Q. Trichosanthin suppresses the proliferation of glioma cells by inhibiting LGR5 expression and the Wnt/β-catenin signaling pathway. Oncol. Rep. 2015, 34, 2845–2852. [Google Scholar] [CrossRef] [Green Version]
- Sha, O.; Yew, D.T.; Cho, E.Y.; Ng, T.B.; Yuan, L.; Kwong, W.H. Mechanism of the specific neuronal toxicity of a type I ribosome-inactivating protein, trichosanthin. Neurotox. Res. 2010, 18, 161–172. [Google Scholar] [CrossRef]
- Cimini, A.; Mei, S.; Benedetti, E.; Laurenti, G.; Koutris, I.; Cinque, B.; Cifone, M.G.; Galzio, R.; Pitari, G.; Di Leandro, L.; et al. Distinct cellular responses induced by saporin and a transferrin-saporin conjugate in two different human glioblastoma cell lines. J. Cell Physiol. 2012, 227, 939–951. [Google Scholar] [CrossRef]
- Bolshakov, A.P.; Stepanichev, M.Y.; Dobryakova, Y.V.; Spivak, Y.S.; Markevich, V.A. Saporin from Saponaria officinalis as a Tool for Experimental Research, Modeling, and Therapy in Neuroscience. Toxins 2020, 12, 546. [Google Scholar] [CrossRef]
- Di Maro, A.; Pizzo, E.; Girbes, T. Biotechnological Potential of Ribosome-Inactivating Proteins (RIPs). In Plant Toxins; Carlini, C.R., Ligabue-Braun, R., Gopalakrishnakone, P., Eds.; Springer: Dordrecht, Netherlands, 2017; pp. 363–381. [Google Scholar]
- Lah Turnšek, T.; Jiao, X.; Novak, M.; Jammula, S.; Cicero, G.; Ashton, A.W.; Joyce, D.; Pestell, R.G. An Update on Glioblastoma Biology, Genetics, and Current Therapies: Novel Inhibitors of the G Protein-Coupled Receptor CCR5. Int. J. Mol. Sci. 2021, 22, 4464. [Google Scholar] [CrossRef]
- Landi, N.; Ruocco, M.R.; Ragucci, S.; Aliotta, F.; Nasso, R.; Pedone, P.V.; Di Maro, A. Quinoa as source of type 1 ribosome inactivating proteins: A novel knowledge for a revision of its consumption. Food Chem. 2021, 342, 128337. [Google Scholar] [CrossRef]
- Santanché, S.; Bellelli, A.; Brunori, M. The unusual stability of saporin, a candidate for the synthesis of immunotoxins. Biochem. Biophys. Res. Commun. 1997, 234, 129–132. [Google Scholar] [CrossRef]
- Tamburino, R.; Pizzo, E.; Sarcinelli, C.; Poerio, E.; Tedeschi, F.; Ficca, A.G.; Parente, A.; Di Maro, A. Enhanced cytotoxic activity of a bifunctional chimeric protein containing a type 1 ribosome-inactivating protein and a serine protease inhibitor. Biochimie 2012, 94, 1990–1996. [Google Scholar] [CrossRef] [PubMed]
- Hanif, F.; Muzaffar, K.; Perveen, K.; Malhi, S.M.; Simjee Sh, U. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac. J. Cancer Prev. APJCP 2017, 18, 3–9. [Google Scholar] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Omar, A.I.; Mason, W.P. Temozolomide: The evidence for its therapeutic efficacy in malignant astrocytomas. Core Evid. 2010, 4, 93–111. [Google Scholar]
- Oliva, M.A.; Staffieri, S.; Castaldo, S.; Giangaspero, F.; Esposito, V.; Arcella, A. Characterization of primary glioma cell lines derived from the patients according to 2016 CNS tumour WHO classification and comparison with their parental tumours. J. Neurooncol. 2021, 151, 123–133. [Google Scholar] [CrossRef]
- Ragucci, S.; Bulgari, D.; Landi, N.; Russo, R.; Clemente, A.; Valletta, M.; Chambery, A.; Gobbi, E.; Faoro, F.; Di Maro, A. The Structural Characterization and Antipathogenic Activities of Quinoin, a Type 1 Ribosome-Inactivating Protein from Quinoa Seeds. Int. J. Mol. Sci. 2021, 22, 8964. [Google Scholar] [CrossRef]
- Di Maro, A.; Valbonesi, P.; Bolognesi, A.; Stirpe, F.; De Luca, P.; Siniscalco Gigliano, G.; Gaudio, L.; Delli Bovi, P.; Ferranti, P.; Malorni, A.; et al. Isolation and characterization of four type-1 ribosome-inactivating proteins, with polynucleotide:adenosine glycosidase activity, from leaves of Phytolacca dioica L. Planta 1999, 208, 125–131. [Google Scholar] [CrossRef]
- Adwan, H.; Murtaja, A.; Kadhim Al-Taee, K.; Pervaiz, A.; Hielscher, T.; Berger, M.R. Riproximin’s activity depends on gene expression and sensitizes PDAC cells to TRAIL. Cancer Biol. Ther. 2014, 15, 1185–1197. [Google Scholar] [CrossRef] [Green Version]
- Ciceroni, C.; Bonelli, M.; Mastrantoni, E.; Niccolini, C.; Laurenza, M.; Larocca, L.M.; Pallini, R.; Traficante, A.; Spinsanti, P.; Ricci-Vitiani, L.; et al. Type-3 metabotropic glutamate receptors regulate chemoresistance in glioma stem cells, and their levels are inversely related to survival in patients with malignant gliomas. Cell Death Differ. 2013, 20, 396–407. [Google Scholar] [CrossRef] [PubMed]
- Surawicz, T.S.; Davis, F.; Freels, S.; Laws, E.R., Jr.; Menck, H.R. Brain tumor survival: Results from the National Cancer Data Base. J. Neurooncol. 1998, 40, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Huszthy, P.C.; Daphu, I.; Niclou, S.P.; Stieber, D.; Nigro, J.M.; Sakariassen, P.O.; Miletic, H.; Thorsen, F.; Bjerkvig, R. In Vivo models of primary brain tumors: Pitfalls and perspectives. Neuro Oncol. 2012, 14, 979–993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahesparan, R.; Read, T.A.; Lund-Johansen, M.; Skaftnesmo, K.O.; Bjerkvig, R.; Engebraaten, O. Expression of extracellular matrix components in a highly infiltrative in vivo glioma model. Acta Neuropathol. 2003, 105, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Fabbrini, M.S.; Katayama, M.; Nakase, I.; Vago, R. Plant Ribosome-Inactivating Proteins: Progesses, Challenges and Biotechnological Applications (and a Few Digressions). Toxins 2017, 9, 314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenblum, M.G.; Cheung, L.H.; Liu, Y.; Marks, J.W., 3rd. Design, expression, purification, and characterization, In Vitro and in vivo, of an antimelanoma single-chain Fv antibody fused to the toxin gelonin. Cancer Res. 2003, 63, 3995–4002. [Google Scholar]
- Shang, C.; Chen, Q.; Dell, A.; Haslam, S.M.; De Vos, W.H.; Van Damme, E.J. The Cytotoxicity of Elderberry Ribosome-Inactivating Proteins Is Not Solely Determined by Their Protein Translation Inhibition Activity. PLoS ONE 2015, 10, e0132389. [Google Scholar] [CrossRef] [Green Version]
- Narayanan, S.; Surendranath, K.; Bora, N.; Surolia, A.; Karande, A.A. Ribosome inactivating proteins and apoptosis. FEBS Lett. 2005, 579, 1324–1331. [Google Scholar] [CrossRef] [Green Version]
- Haar, C.P.; Hebbar, P.; Wallace, G.C.t.; Das, A.; Vandergrift, W.A., 3rd.; Smith, J.A.; Giglio, P.; Patel, S.J.; Ray, S.K.; Banik, N.L. Drug resistance in glioblastoma: A mini review. Neurochem. Res. 2012, 37, 1192–1200. [Google Scholar] [CrossRef]
- Schneider, S.W.; Ludwig, T.; Tatenhorst, L.; Braune, S.; Oberleithner, H.; Senner, V.; Paulus, W. Glioblastoma cells release factors that disrupt blood-brain barrier features. Acta Neuropathol. 2004, 107, 272–276. [Google Scholar] [CrossRef]
- Sarkaria, J.N.; Hu, L.S.; Parney, I.F.; Pafundi, D.H.; Brinkmann, D.H.; Laack, N.N.; Giannini, C.; Burns, T.C.; Kizilbash, S.H.; Laramy, J.K.; et al. Is the blood-brain barrier really disrupted in all glioblastomas? A critical assessment of existing clinical data. Neuro Oncol. 2018, 20, 184–191. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rotondo, R.; Ragucci, S.; Castaldo, S.; Oliva, M.A.; Landi, N.; Pedone, P.V.; Arcella, A.; Di Maro, A. Cytotoxicity Effect of Quinoin, Type 1 Ribosome-Inactivating Protein from Quinoa Seeds, on Glioblastoma Cells. Toxins 2021, 13, 684. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13100684
Rotondo R, Ragucci S, Castaldo S, Oliva MA, Landi N, Pedone PV, Arcella A, Di Maro A. Cytotoxicity Effect of Quinoin, Type 1 Ribosome-Inactivating Protein from Quinoa Seeds, on Glioblastoma Cells. Toxins. 2021; 13(10):684. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13100684
Chicago/Turabian StyleRotondo, Rossella, Sara Ragucci, Salvatore Castaldo, Maria Antonietta Oliva, Nicola Landi, Paolo V. Pedone, Antonietta Arcella, and Antimo Di Maro. 2021. "Cytotoxicity Effect of Quinoin, Type 1 Ribosome-Inactivating Protein from Quinoa Seeds, on Glioblastoma Cells" Toxins 13, no. 10: 684. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13100684