Salinity Affects Saxitoxins (STXs) Toxicity in the Dinoflagellate Alexandrium pacificum, with Low Transcription of SXT-Biosynthesis Genes sxtA4 and sxtG
Abstract
:1. Introduction
2. Results
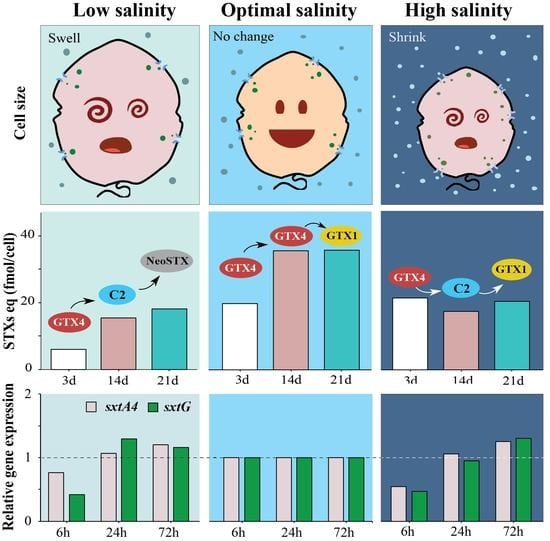
2.1. Effects of Salinity on Cell Growth and Cell Size
2.2. Toxin Content and Composition at Different Salinities
2.3. Expression Levels of sxtA4 and sxtG in A. pacificum under Different Salinities
2.4. Statistical Correlations among Salinity Concentrations, Gene Expression, and Biological Data
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Strain and Culture Conditions
5.2. Salinity Experiment Setup
5.3. Cell Growth and Cell Size Measurements
5.4. STX Analysis
5.5. RNA Extraction and cDNA Synthesis
5.6. Quantitative Real-Time PCR
5.7. Statistical and Principal Component Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vandersea, M.W.; Kibler, S.R.; Tester, P.A.; Holderied, K.; Hondolero, D.E.; Powell, K.; Baird, S.; Doroff, A.; Dugan, D.; Litaker, R.W. Environmental factors influencing the distribution and abundance of Alexandrium catenella in Kachemak bay and lower cook inlet, Alaska. Harmful Algae 2018, 77, 81–92. [Google Scholar] [CrossRef]
- Condie, S.A.; Oliver, E.C.J.; Hallegraeff, G.M. Environmental drivers of unprecedented Alexandrium catenella dinoflagellate blooms off eastern Tasmania, 2012–2018. Harmful Algae 2019, 87, 101628. [Google Scholar] [CrossRef]
- Algaebase. World-Wide Electronic Publication, National University of Ireland, Galway. Available online: http://www.algaebase.org (accessed on 20 June 2021).
- Anderson, D.M.; Alpermann, T.J.; Cembella, A.D.; Collos, Y.; Masseret, E.; Montresor, M. The globally distributed genus Alexandrium: Multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae 2012, 14, 10–35. [Google Scholar] [CrossRef] [Green Version]
- Caruana, A.M.N.; Amzil, Z. Microalgae and toxins. In Microalgae in Health and Disease Prevention, 1st ed.; Levine, I.A., Fleurence, J., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 263–305. [Google Scholar]
- Blossom, H.E.; Markussen, B.; Daugbjerg, N.; Krock, B.; Norlin, A.; Hansen, P.J. The cost of toxicity in microalgae: Direct evidence from the dinoflagellate Alexandrium. Front. Microbiol. 2019, 10, 1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bricelj, V.M.; Shumway, S.E. Paralytic shellfish toxins in bivalve molluscs: Occurrence, transfer kinetics, and biotransformation. Rev. Fish. Sci. 1998, 6, 315–383. [Google Scholar] [CrossRef]
- Brown, A.R.; Lilley, M.; Shutler, J.; Lowe, C.; Artioli, Y.; Torres, R.; Berdalet, E.; Tyler, C.R. Assessing risks and mitigating impacts of harmful algal blooms on mariculture and marine fisheries. Rev. Aquac. 2020, 12, 1663–1688. [Google Scholar] [CrossRef] [Green Version]
- Harmful Algae Event Database (HAEDAT). Available online: http://haedat.iode.org (accessed on 21 June 2021).
- Schantz, E.J.; Mold, J.D.; Stanger, D.W.; Shavel, J.; Riel, F.J.; Bowden, J.P.; Lynch, J.M.; Wyler, R.S.; Riegel, B.; Sommer, H. Paralytic shellfish poison. VI. A procedure for the isolation and purification of the poison from toxic clam and mussel tissues. J. Am. Chem. Soc. 1957, 79, 5230–5235. [Google Scholar] [CrossRef]
- Oshima, Y.; Blackburn, S.I.; Hallegraeff, G.M. Comparative study on paralytic shellfish toxin profiles of the dinoflagellate Gymnodinium catenatum from three different countries. Mar. Biol. Int. J. Life Ocean. Coast. Waters 1993, 116, 471–476. [Google Scholar] [CrossRef]
- Usup, G.; Kulis, D.M.; Anderson, D.M. Growth and toxin production of the toxic dinoflagellate Pyrodinium var. compressum in laboratory cultures. Nat. Toxins 1994, 2, 254–262. [Google Scholar] [CrossRef]
- Stüken, A.; Orr, R.J.S.; Kellmann, R.; Murray, S.A.; Neilan, B.A.; Jakobsen, K.S. Discovery of nuclear-encoded genes for the neurotoxin saxitoxin in dinoflagellates. PLoS ONE 2011, 6, e20096. [Google Scholar] [CrossRef] [Green Version]
- Kellmann, R.; Neilan, B.A. Biochemical characterization of paralytic shellfish toxin biosynthesis in vitro. J. Phycol. 2007, 43, 497–508. [Google Scholar] [CrossRef]
- Wiese, M.; D’Agostino, P.M.; Mihali, T.K.; Moffitt, M.C.; Neilan, B.A. Neurotoxic alkaloids: Saxitoxin and its analogs. Mar. Drugs 2010, 8, 2185–2211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ongley, S.E.; Pengelly, J.J.L.; Neilan, B.A. Elevated Na+ and pH influence the production and transport of saxitoxin in the cyanobacteria Anabaena circinalis AWQC131C and Cylindrospermopsis raciborskii T3. Environ. Microbiol. 2016, 18, 427–438. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, P.; Moffitt, M.; Neilan, B. Current knowledge of paralytic shellfish toxin biosynthesis, molecular detection and evolution. In Toxins and Biologically Active Compounds from Microalgae: Volume I, Origin, Chemistry and Detection, 1st ed.; Rossini, G.P., Ed.; CRC Press: Boca Raton, FL, USA, 2014; pp. 251–280. [Google Scholar]
- Wang, D.Z.; Zhang, S.F.; Zhang, Y.; Lin, L. Paralytic shellfish toxin biosynthesis in cyanobacteria and dinoflagellates: A molecular overview. J. Proteomics 2016, 135, 132–140. [Google Scholar] [CrossRef]
- Walker, J.R.; Merit, J.E.; Thomas-Tran, R.; Tang, D.T.Y.; Bois, D.J. Divergent synthesis of natural derivatives of (+)-Saxitoxin including 11-Saxitoxinethanoic acid. Angew. Chem. 2019, 131, 1703–1707. [Google Scholar] [CrossRef]
- Anderson, D.M.; Glibert, P.M.; Burkholder, J.M. Harmful algal blooms and eutrophication: Nutrient sources, composition, and consequences. Estuaries 2002, 25, 704–726. [Google Scholar] [CrossRef]
- Etheridge, S.M.; Roesler, C.S. Effects of temperature, irradiance, and salinity on photosynthesis, growth rates, total toxicity, and toxin composition for Alexandrium fundyense isolates from the Gulf of Maine and Bay of Fundy. Deep. Res. Part. II Top. Stud. Oceanogr. 2005, 52, 2491–2500. [Google Scholar] [CrossRef]
- Grzebyk, D.; Béchemin, C.; Ward, C.J.; Vérité, C.; Codd, G.A.; Maestrini, S.Y. Effects of salinity and two coastal waters on the growth and toxin content of the dinoflagellate Alexandrium minutum. J. Plankton Res. 2003, 25, 1185–1199. [Google Scholar] [CrossRef]
- Parkhill, J.P.; Cembella, A.D. Effects of salinity, light and inorganic nitrogen on growth and toxigenicity of the marine dinoflagellate Alexandrium tamarense from northeastern Canada. J. Plankton Res. 1999, 21, 939–955. [Google Scholar] [CrossRef]
- Wang, H.; Kim, H.; Park, H.; Ki, J.S. Temperature influences the content and biosynthesis gene expression of saxitoxins (STXs) in the toxigenic dinoflagellate Alexandrium pacificum. Sci. Total Environ. 2021, 802, 149801. [Google Scholar] [CrossRef]
- Hii, K.S.; Lim, P.T.; Kon, N.F.; Takata, Y.; Usup, G.; Leaw, C.P. Physiological and transcriptional responses to inorganic nutrition in a tropical Pacific strain of Alexandrium minutum: Implications for the saxitoxin genes and toxin production. Harmful Algae 2016, 56, 9–21. [Google Scholar] [CrossRef]
- Mesquita, M.C.B.; Lürling, M.; Dorr, F.; Pinto, E.; Marinho, M.M. Combined effect of light and temperature on the production of saxitoxins in Cylindrospermopsis raciborskii strains. Toxins 2019, 11, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alkawri, A.A.S.; Ramaiah, N. Spatio-temporal variability of dinoflagellate assemblages in different salinity regimes in the west coast of India. Harmful Algae 2010, 9, 153–162. [Google Scholar] [CrossRef]
- Kirst, G.O. Salinity tolerance of eukaryotic marine algae. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 1990, 41, 21–53. [Google Scholar] [CrossRef]
- Errera, R.M.; Campbell, L. Osmotic stress triggers toxin production by the dinoflagellate Karenia brevis. Proc. Natl. Acad. Sci. USA 2012, 108, 10597–10601. [Google Scholar] [CrossRef] [Green Version]
- Laabir, M.; Collos, Y.; Masseret, E.; Grzebyk, D.; Abadie, E.; Savar, V.; Sibat, M.; Amzil, Z. Influence of environmental factors on the paralytic shellfish toxin content and profile of Alexandrium catenella (Dinophyceae) isolated from the Mediterranean Sea. Mar. Drugs 2013, 11, 1583–1601. [Google Scholar] [CrossRef] [Green Version]
- Lim, P.T.; Ogata, T. Salinity effect on growth and toxin production of four tropical Alexandrium species (Dinophyceae). Toxicon 2005, 45, 699–710. [Google Scholar] [CrossRef]
- Shimizu, Y.; Norte, M.; Hori, A.; Genenah, A.; Kobayashi, M. Biosynthesis of saxitoxin analogs: The unexpected pathway. J. Am. Chem. Soc. 1984, 106, 6433–6434. [Google Scholar] [CrossRef]
- Kellmann, R.; Mihali, T.K.; Young, J.J.; Pickford, R.; Pomati, F.; Neilan, B.A. Biosynthetic intermediate analysis and functional homology reveal a saxitoxin gene cluster in cyanobacteria. Appl. Environ. Microbiol. 2008, 74, 4044–4053. [Google Scholar] [CrossRef] [Green Version]
- Verma, A.; Barua, A.; Ruvindy, R.; Savela, H.; Ajani, P.A.; Murray, S.A. The genetic basis of toxin biosynthesis in dinoflagellates. Microorganisms 2019, 7, 222. [Google Scholar] [CrossRef] [Green Version]
- Pearson, L.; Mihali, T.; Moffitt, M.; Kellmann, R.; Neilan, B. On the chemistry, toxicology and genetics of the cyanobacterial toxins, microcystin, nodularin, saxitoxin and cylindrospermopsin. Marine Drugs 2010, 8, 1650–1680. [Google Scholar] [CrossRef] [Green Version]
- Murray, S.A.; Diwan, R.; Orr, R.J.S.; Kohli, G.S.; John, U. Gene duplication, loss and selection in the evolution of saxitoxin biosynthesis in alveolates. Mol. Phylogenet. Evol. 2015, 92, 165–180. [Google Scholar] [CrossRef] [Green Version]
- Suikkanen, S.; Kremp, A.; Hautala, H.; Krock, B. Paralytic shellfish toxins or spirolides? The role of environmental and genetic factors in toxin production of the Alexandrium ostenfeldii complex. Harmful Algae 2013, 26, 52–59. [Google Scholar] [CrossRef]
- Wang, H.; Kim, H.; Ki, J.S. Transcriptome survey and toxin measurements reveal evolutionary modification and loss of saxitoxin biosynthesis genes in the dinoflagellates Amphidinium carterae and Prorocentrum micans. Ecotoxicol. Environ. Saf. 2020, 195, 110474. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.F.; Lin, L.; Wang, D.Z. Comparative transcriptome analysis of a toxin-producing dinoflagellate Alexandrium catenella and its non-toxic mutant. Mar. Drugs 2014, 12, 5698–5718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, S.A.; Hoppenrath, M.; Orr, R.J.S.; Bolch, C.; John, U.; Diwan, R.; Yauwenas, R.; Harwood, T.; Salas, D.M.; Neilan, B.; et al. Alexandrium diversaporum sp. nov., a new non-saxitoxin producing species: Phylogeny, morphology and sxtA genes. Harmful Algae 2014, 31, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Perini, F.; Galluzzi, L.; Dell’Aversano, C.; Iacovo, E.D.; Tartaglione, L.; Ricci, F.; Forino, M.; Ciminiello, P.; Penna, A. SxtA and sxtG gene expression and toxin production in the mediterranean Alexandrium minutum (Dinophyceae). Mar. Drugs 2014, 12, 5258–5276. [Google Scholar] [CrossRef]
- Kim, H.; Park, H.; Wang, H.; Yoo, H.Y.; Park, J.; Ki, J. Low temperature and cold stress significantly increase saxitoxins (STXs) and expression of STX biosynthesis genes sxtA4 and sxtG in the dinoflagellate Alexandrium catenella. Mar. Drugs 2021, 19, 291. [Google Scholar] [CrossRef]
- Fertouna-Bellakhal, M.; Dhib, A.; Fathalli, A.; Bellakhal, M.; Chomérat, N.; Masseret, E.; Laabir, M.; Turki, S.; Aleya, L. Alexandrium pacificum Litaker sp. nov (Group IV): Resting cyst distribution and toxin profile of vegetative cells in Bizerte Lagoon (Tunisia, Southern Mediterranean Sea). Harmful Algae 2015, 48, 69–82. [Google Scholar] [CrossRef]
- Barua, A.; Ajani, P.A.; Ruvindy, R.; Farrell, H.; Zammit, A.; Brett, S.; Hill, D.; Sarowar, C.; Hoppenrath, M.; Murray, S.A. First detection of paralytic shellfish toxins from Alexandrium pacificum above the regulatory limit in blue mussels (Mytilus galloprovincialis) in New South Wales, Australia. Microorganisms 2020, 8, 905. [Google Scholar] [CrossRef]
- Nagai, S.; Matsuyama, Y.; Oh, S.J.; Itakura, S. Effect of nutrients and temperature on encystment of the toxic dinoflagellate Alexandrium tamarense (Dinophyceae) isolated from Hiroshima Bay, Japan. Plankt. Biol. Ecol. 2004, 51, 103–109. [Google Scholar]
- Dai, L.; Yu, R.C.; Geng, H.X.; Zhao, Y.; Zhang, Q.C.; Kong, F.Z.; Chen, Z.F.; Zhao, J.Y.; Zhou, M.J. Resting cysts of Alexandrium catenella and A. pacificum (Dinophyceae) in the Bohai and Yellow Seas, China: Abundance, distribution and implications for toxic algal blooms. Harmful Algae 2020, 93, 101794. [Google Scholar] [CrossRef] [PubMed]
- Hadjadji, I.; Laabir, M.; Frihi, H.; Collos, Y.; Shao, Z.J.; Berrebi, P.; Abadie, E.; Amzil, Z.; Chomérat, N.; Rolland, J.L.; et al. Unsuspected intraspecific variability in the toxin production, growth and morphology of the dinoflagellate Alexandrium pacificum R.W. Litaker (Group IV) blooming in a South Western Mediterranean marine ecosystem, Annaba Bay (Algeria). Toxicon 2020, 180, 79–88. [Google Scholar] [CrossRef]
- Han, M.S.; Jeon, J.K.; Kim, Y.O. Occurrence of dinoflagellate Alexandrium tamarense, a causative organism of paralytic shellfish poisoning in Chinhae Bay, Korea. J. Plankton Res. 1992, 14, 1581–1592. [Google Scholar] [CrossRef]
- Kim, Y.O.; Park, M.H.; Han, M.S. Role of cyst germination in the bloom initiation of Alexandrium tamarense (Dinophyceae) in Masan Bay, Korea. Aquat. Microb. Ecol. 2002, 29, 279–286. [Google Scholar] [CrossRef]
- Lee, H.G.; Kim, H.M.; Min, J.; Park, C.; Jeong, H.J.; Lee, K.; Kim, K.Y. Quantification of the paralytic shellfish poisoning dinoflagellate Alexandrium species using a digital PCR. Harmful Algae 2020, 92, 101726. [Google Scholar] [CrossRef] [PubMed]
- John, U.; Litaker, R.W.; Montresor, M.; Murray, S.; Brosnahan, M.L.; Anderson, D.M. Formal revision of the Alexandrium tamarense species complex (dinophyceae) taxonomy: The introduction of five species with emphasis on molecular-based (rDNA) classification. Protist 2014, 165, 779–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bill, B.D.; Moore, S.K.; Hay, L.R.; Anderson, D.M.; Trainer, V.L. Effects of temperature and salinity on the growth of Alexandrium (Dinophyceae) isolates from the Salish Sea. J. Phycol. 2016, 52, 230–238. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Kim, H.; Ki, J.S. Preliminary result of de novo transcriptome sequencing of the marine toxic dinoflagellate Alexandrium catenella incubated under several different stresses. Mar. Biol. 2021, 168, 1–8. [Google Scholar] [CrossRef]
- Antonella, P.; Luca, G. The quantitative real-time PCR applications in the monitoring of marine harmful algal bloom (HAB) species. Environ. Sci. Pollut. Res. 2013, 20, 6851–6862. [Google Scholar] [CrossRef] [Green Version]
- Stucken, K.; John, U.; Cembella, A.; Soto-liebe, K. Impact of nitrogen sources on gene expression and toxin production in the diazotroph Cylindrospermopsis raciborskii CS-505 and non-diazotroph Raphidiopsis brookii D9. Toxins 2014, 6, 1896–1915. [Google Scholar] [CrossRef] [Green Version]
- Kremp, A.; Hansen, P.J.; Tillmann, U.; Savela, H.; Suikkanen, S.; Voß, D.; Barrera, F.; Jakobsen, H.H.; Krock, B. Distributions of three Alexandrium species and their toxins across a salinity gradient suggest an increasing impact of GDA producing A. pseudogonyaulax in shallow brackish waters of Northern Europe. Harmful Algae 2019, 87, 101622. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.T.; Leaw, C.P.; Sato, S.; Van Thuoc, C.; Kobiyama, A.; Ogata, T. Effect of salinity on growth and toxin production of Alexandrium minutum isolated from a shrimp culture pond in northern Vietnam. J. Appl. Phycol. 2011, 23, 857–864. [Google Scholar] [CrossRef] [Green Version]
- Oh, S.J.; Park, J.A.; Kwon, H.K.; Yang, H.S.; Lim, W.A. Ecophysiological studies on the population dynamics of two toxic dinoflagellates Alexandrium tamarense and Alexandrium catenella isolated from the Southern Coast of Korea-I. Effects of temperature and salinity on the growth. J. Korean Soc. Mar. Environ. Energy 2012, 15, 133–141. [Google Scholar] [CrossRef]
- Shin, H.H.; Baek, S.H.; Li, Z.; Han, M.S.; Oh, S.J.; Youn, S.H.; Kim, Y.S.; Kim, D.; Lim, W.A. Resting cysts, and effects of temperature and salinity on the growth of vegetative cells of the potentially harmful species Alexandrium insuetum Balech (Dinophyceae). Harmful Algae 2014, 39, 175–184. [Google Scholar] [CrossRef]
- Lim, P.T.; Leaw, C.P.; Usup, G.; Kobiyama, A.; Koike, K.; Ogata, T. Effects of light and temperature on growth, nitrate uptake, and toxin production of two tropical dinoflagellates: Alexandrium tamiyavanichii and Alexandrium minutum (Dinophyceae). J. Phycol. 2006, 42, 786–799. [Google Scholar] [CrossRef]
- Aguilera-Belmonte, A.; Inostroza, I.; Carrillo, K.S.; Franco, J.M.; Riobó, P.; Gómez, P.I. The combined effect of salinity and temperature on the growth and toxin content of four Chilean strains of Alexandrium catenella (Whedon and Kofoid) Balech 1985 (Dinophyceae) isolated from an outbreak occurring in southern Chile in 2009. Harmful Algae 2013, 13, 55–59. [Google Scholar] [CrossRef]
- Maclean, C.; Cembella, A.D.; Quilliam, M.A. Effects of light, salinity and inorganic nitrogen on cell growth and spirolide production in the marine dinoflagellate Alexandrium ostenfeldii (Paulsen) Balech et Tangen. Bot. Mar. 2003, 46, 466–476. [Google Scholar] [CrossRef]
- Martens, H.; Waal, D.B.V.D.; Brandenburg, K.M.; Krock, B.; Tillmann, U. Salinity effects on growth and toxin production in an Alexandrium ostenfeldii (Dinophyceae) isolate from The Netherlands. J. Plankton Res. 2016, 38, 1302–1316. [Google Scholar] [CrossRef] [Green Version]
- Olenina, I.; Vaičiukynas, E.; Šulčius, S.; Paškauskas, R.; Verikas, A.; Gelžinis, A.; Bačauskiene, M.; Bertašiute, V.; Olenin, S. The dinoflagellate Prorocentrum cordatum at the edge of the salinity tolerance: The growth is slower but cells are larger. Estuar. Coast. Shelf Sci. 2016, 168, 71–79. [Google Scholar] [CrossRef]
- Mayfield, A.B.; Gates, R.D. Osmoregulation in anthozoan-dinoflagellate symbiosis. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2007, 147, 1–10. [Google Scholar] [CrossRef]
- Jensen, M.Ø.; Moestrup, Ø. Autecology of the toxic dinoflagellate Alexandrium ostenfeldii: Life history and growth at different temperatures and salinities. Eur. J. Phycol. 1997, 32, 9–18. [Google Scholar] [CrossRef]
- Kasinathan, V.; Wingler, A. Effect of reduced arginine decarboxylase activity on salt tolerance and on polyamine formation during salt stress in Arabidopsis thaliana. Physiol. Plant. 2004, 121, 101–107. [Google Scholar] [CrossRef]
- Lazcano-Ferrat, I.; Lovatt, C.J. Effect of salinity on arginine biosynthesis in leaves of Phaseolus vulgaris L. and P. acutifolius A. Gray. Crop. Sci. 1997, 37, 469–475. [Google Scholar] [CrossRef]
- Munday, R.; Thomas, K.; Gibbs, R.; Murphy, C.; Quilliam, M.A. Acute toxicities of saxitoxin, neosaxitoxin, decarbamoyl saxitoxin and gonyautoxins 1&4 and 2&3 to mice by various routes of administration. Toxicon 2013, 76, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Selwood, A.I.; Waugh, C.; Harwood, D.T.; Rhodes, L.L.; Reeve, J.; Sim, J.; Munday, R. Acute toxicities of the saxitoxin congeners gonyautoxin 5, gonyautoxin 6, decarbamoyl gonyautoxin 2&3, decarbamoyl neosaxitoxin, C-1&2 and C-3&4 to mice by various routes of administration. Toxins 2017, 9, 73. [Google Scholar] [CrossRef]
- Wang, H.; Guo, R.; Lim, W.A.; Allen, A.E.; Ki, J.S. Comparative transcriptomics of toxin synthesis genes between the non-toxin producing dinoflagellate Cochlodinium polykrikoides and toxigenic Alexandrium pacificum. Harmful Algae 2020, 93, 101777. [Google Scholar] [CrossRef]
- Cirés, S.; Delgado, A.; González-Pleiter, M.; Quesada, A. Temperature influences the production and transport of saxitoxin and the expression of sxt genes in the cyanobacterium Aphanizomenon gracile. Toxins 2017, 9, 322. [Google Scholar] [CrossRef] [Green Version]
- Guillard, R.R.L. Culture of phytoplankton for feeding marine invertebrates. In Culture of Marine Invertebrate Animal, 1st ed.; Chanley, M.H., Smith, W.L., Eds.; Plenum Press: New York, NY, USA, 1975; pp. 29–60. [Google Scholar]
- Kim, J.H.; Oh, S.J.; Kim, S.-Y. The effect of temperature, salinity and irradiance on the growth of Alexandrium affine (Dinophyceae) isolated from southern sea of Korea. J. Korean Soc. Mar. Environ. Saf. 2019, 25, 229–236. [Google Scholar] [CrossRef]
- Nam, K.T.; Oh, S.J. Influence of water temperature and salinity on the production of paralytic shellfish poisoning by toxic dinoflagellate Alexandrium catenella (Group I). J. Korean Soc. Mar. Environ. Saf. 2021, 27, 119–126. [Google Scholar] [CrossRef]
- Yamamoto, T.; Oh, S.J.; Kataoka, Y. Effects of water temperature, salinity and irradiance on the growth of the toxic dinoflagellate, Gymnodinium catenatum (Graham) isolated from Hiroshima Bay, Japan. Fish. Sci. 2002, 68, 356–363. [Google Scholar] [CrossRef] [Green Version]
- Rey, V.; Botana, A.M.; Alvarez, M.; Antelo, A.; Botana, L.M. Liquid chromatography with a fluorimetric detection method for analysis of paralytic shellfish toxins and tetrodotoxin based on a porous graphitic carbon column. Toxins 2016, 8, 196. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T. Morphometrics. In Paleontological Data Analysis; Wiley: Hoboken, NJ, USA, 2008; pp. 78–156. [Google Scholar]





| Species | Strain | Salinity Range | Optimal Growth Salinity | Highest Toxin Condition | Toxins | STXs Eq (fmol/cell) | Reference |
|---|---|---|---|---|---|---|---|
| A. catenella | PFB38 | 15–35 | 35 | 35 | NeoSTX, GTX1-5 | 95.76 | [61] |
| ACT03 | 10–40 | 30 | 35 | C1-4, GTX3-5 | 50.3 | [30] | |
| A. fundyense | MI | 15–35 | 25 | 30 | STX, NeoSTX, GTX1-4 | 62 | [21] |
| BoF | 15–35 | 25 | 30–35 | STX, NeoSTX, GTX1-4 | 73–75 | [21] | |
| A. minutum | AM89BM | 12–37 | 20–37 | 15 | - | 50 | [22] |
| AmKB06 | 2–30 | 15 | 5 | GTX1-6, C2, NeoSTX, dcSTX | 12 | [31] | |
| Alexsp17 | 5–35 | 10–15 | 30–35 | STX, NeoSTX, dcSTX, C2, GTX2-4, GTX4-12ol | 30 | [57] | |
| A. ostenfeldii | AOSH1 | 15–33 | 33 | 15 | C3, C, desmthyl D | - | [62] |
| OKNL21 | 3–34 | 22 | 5 | STX, GTX2/3/5, C1-2 | 52 | [63] | |
| A. peruvianum | ApKS01 | 2–30 | 25 | 25 | GTX1-6, C2, NeoSTX, dcSTX | 0.75 | [31] |
| A. tamarense | Pr18b | 10–30 | 25‰ | 25 | STX, NeoSTX, GTX1-4, C1-3 | 179 | [23] |
| AtPA01 | 2–30 | 20–30 | 15 | GTX1-6, C2, NeoSTX, dcSTX | 0.8 | [31] | |
| A. tamiyavanichii | AcMS01 | 2–30 | 25 | 20 | GTX1-6, C2, NeoSTX, dcSTX | 80 | [31] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bui, Q.T.N.; Kim, H.; Park, H.; Ki, J.-S. Salinity Affects Saxitoxins (STXs) Toxicity in the Dinoflagellate Alexandrium pacificum, with Low Transcription of SXT-Biosynthesis Genes sxtA4 and sxtG. Toxins 2021, 13, 733. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13100733
Bui QTN, Kim H, Park H, Ki J-S. Salinity Affects Saxitoxins (STXs) Toxicity in the Dinoflagellate Alexandrium pacificum, with Low Transcription of SXT-Biosynthesis Genes sxtA4 and sxtG. Toxins. 2021; 13(10):733. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13100733
Chicago/Turabian StyleBui, Quynh Thi Nhu, Hansol Kim, Hyunjun Park, and Jang-Seu Ki. 2021. "Salinity Affects Saxitoxins (STXs) Toxicity in the Dinoflagellate Alexandrium pacificum, with Low Transcription of SXT-Biosynthesis Genes sxtA4 and sxtG" Toxins 13, no. 10: 733. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13100733