Fungi–Bacteria Correlation in Alcoholic Hepatitis Patients
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
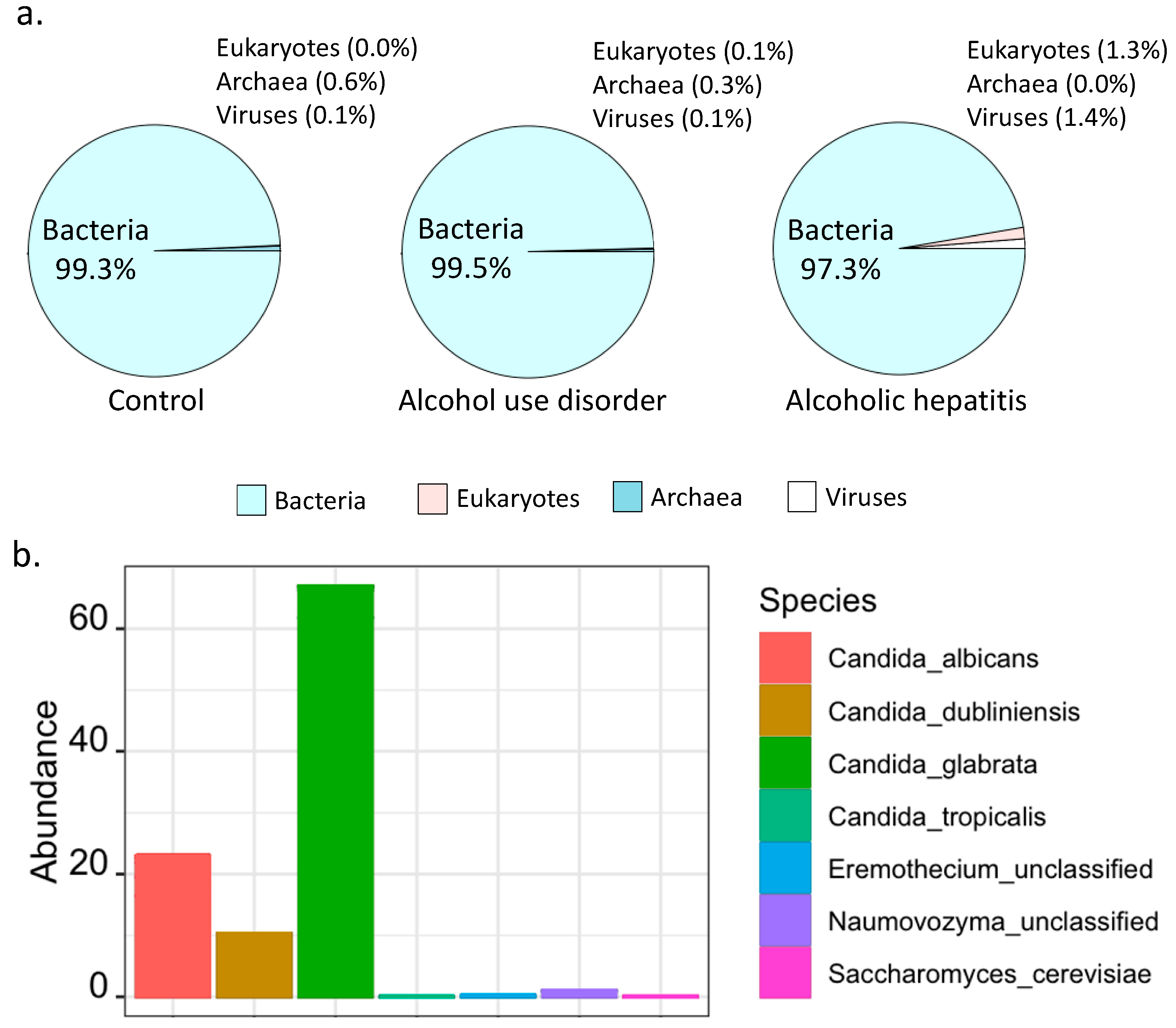
2.2. Percentage of Bacteria and Fungi in Alcoholic Hepatitis Patients
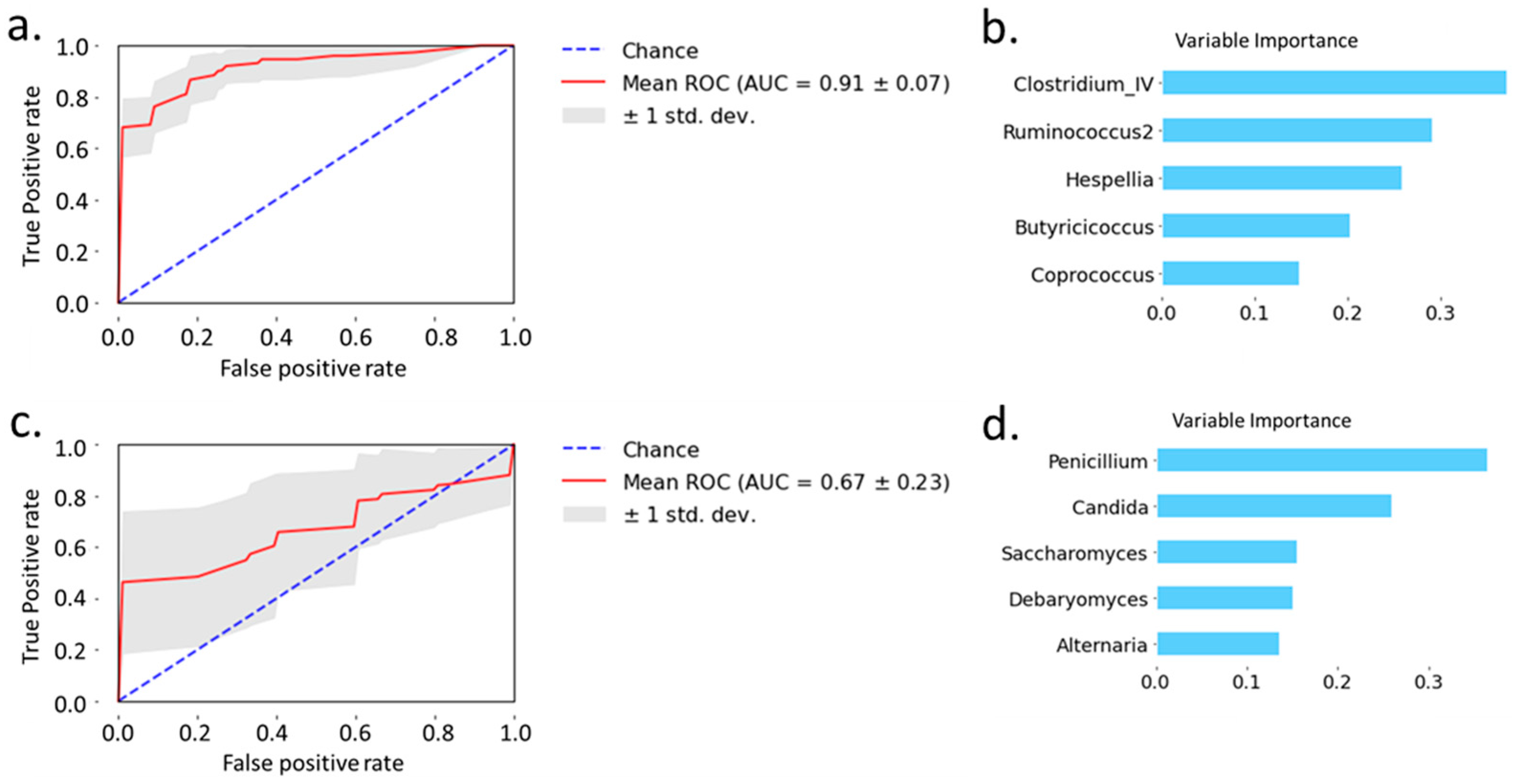
2.3. Prediction of Alcoholic Hepatitis
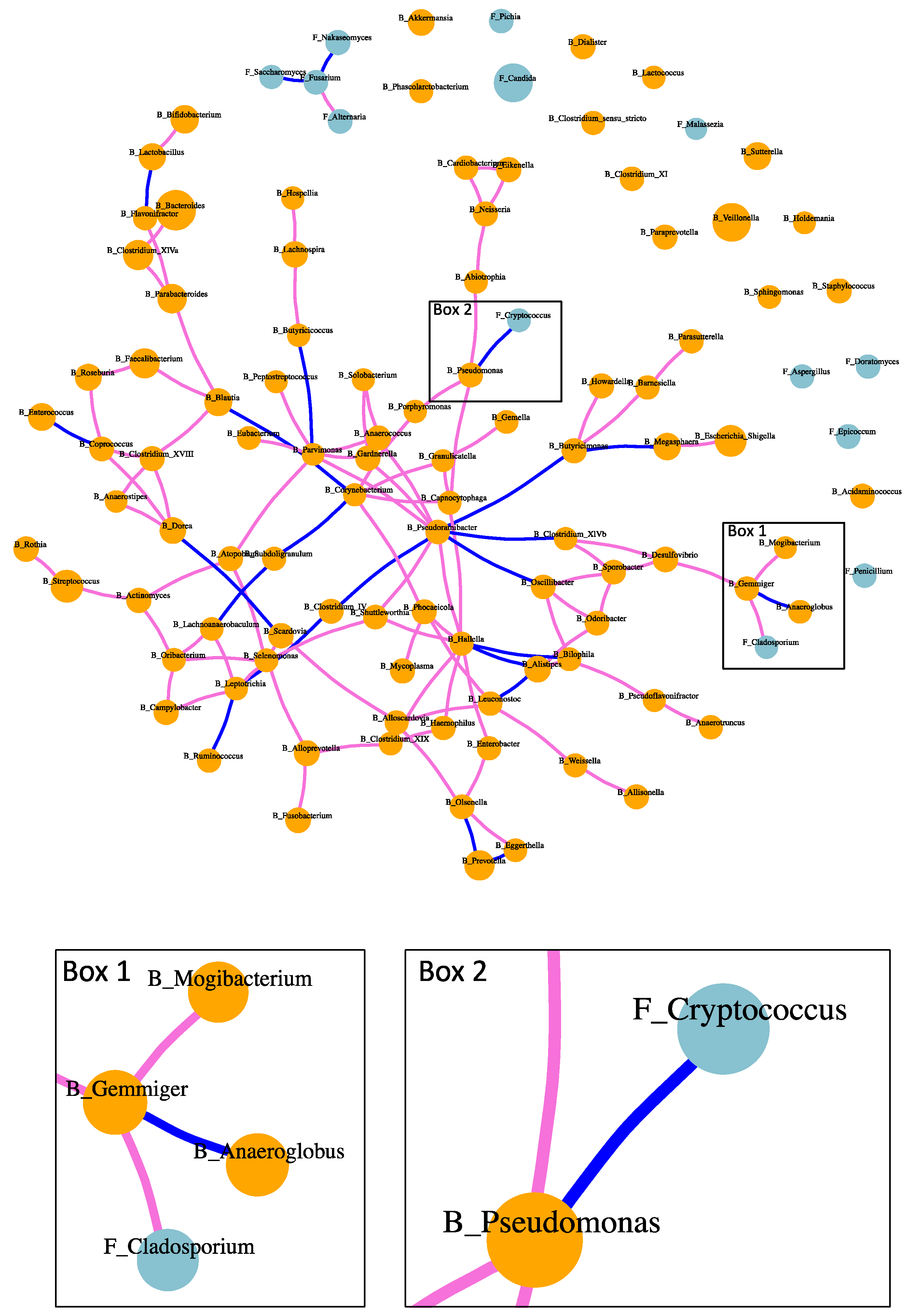
2.4. Fungi–Bacteria Network
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Shotgun Metagenomic Analysis
4.3. ITS Sequencing
4.4. 16S rRNA Sequencing
4.5. Random Forest Model
4.6. Fungi–Bacteria Network in Alcoholic Hepatitis Patients
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thursby, E.; Juge, N. Introduction to the Human Gut Microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Osna, N.A.; Donohue, T.M.; Kharbanda, K.K. Alcoholic Liver Disease: Pathogenesis and Current Management. Alcohol. Res. 2017, 38, 147–161. [Google Scholar]
- Casafont Morencos, F.; de las Heras Castaño, G.; Martín Ramos, L.; López Arias, M.J.; Ledesma, F.; Pons Romero, F. Small Bowel Bacterial Overgrowth in Patients with Alcoholic Cirrhosis. Dig. Dis. Sci. 1996, 41, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Lang, S.; Fairfied, B.; Gao, B.; Duan, Y.; Zhang, X.; Fouts, D.E.; Schnabl, B. Changes in the Fecal Bacterial Microbiota Associated with Disease Severity in Alcoholic Hepatitis Patients. Gut Microbes 2020, 12, 1785251. [Google Scholar] [CrossRef]
- Lang, S.; Duan, Y.; Liu, J.; Torralba, M.G.; Kuelbs, C.; Ventura-Cots, M.; Abraldes, J.G.; Bosques-Padilla, F.; Verna, E.C.; Brown, R.S.; et al. Intestinal Fungal Dysbiosis and Systemic Immune Response to Fungi in Patients with Alcoholic Hepatitis. Hepatology 2020, 71, 522–538. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Coughlin, L.A.; Neubauer, M.M.; Kim, J.; Kim, M.S.; Zhan, X.; Simms-Waldrip, T.R.; Xie, Y.; Hooper, L.V.; Koh, A.Y. Activation of HIF-1α and LL-37 by Commensal Bacteria Inhibits Candida Albicans Colonization. Nat. Med. 2015, 21, 808–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuskin, F.; Lowe, E.C.; Temple, M.J.; Zhu, Y.; Cameron, E.; Pudlo, N.A.; Porter, N.T.; Urs, K.; Thompson, A.J.; Cartmell, A.; et al. Human Gut Bacteroidetes Can Utilize Yeast Mannan through a Selfish Mechanism. Nature 2015, 517, 165–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Everard, A.; Matamoros, S.; Geurts, L.; Delzenne, N.M.; Cani, P.D. Saccharomyces Boulardii Administration Changes Gut Microbiota and Reduces Hepatic Steatosis, Low-Grade Inflammation, and Fat Mass in Obese and Type 2 Diabetic Db/Db Mice. mBio 2014, 5, e01011–e01014. [Google Scholar] [CrossRef] [Green Version]
- Cooper, T.G. Allantoin Degradation by Saccharomyces Cerevisiae--a Model System for Gene Regulation and Metabolic Integration. Adv. Enzymol. Relat. Areas Mol. Biol. 1984, 56, 91–139. [Google Scholar] [CrossRef]
- Wang, M.; Chen, W.-Y.; Zhang, J.; Gobejishvili, L.; Barve, S.S.; McClain, C.J.; Joshi-Barve, S. Elevated Fructose and Uric Acid Through Aldose Reductase Contribute to Experimental and Human Alcoholic Liver Disease. Hepatology 2020, 72, 1617–1637. [Google Scholar] [CrossRef] [PubMed]
- Ben Haj Khalifa, A.; Moissenet, D.; Vu Thien, H.; Khedher, M. Virulence factors in Pseudomonas aeruginosa: Mechanisms and modes of regulation. Ann. Biol. Clin. 2011, 69, 393–403. [Google Scholar] [CrossRef]
- May, R.C.; Stone, N.R.H.; Wiesner, D.L.; Bicanic, T.; Nielsen, K. Cryptococcus: From Environmental Saprophyte to Global Pathogen. Nat. Rev. Microbiol. 2016, 14, 106–117. [Google Scholar] [CrossRef]
- Rella, A.; Yang, M.W.; Gruber, J.; Montagna, M.T.; Luberto, C.; Zhang, Y.-M.; Del Poeta, M. Pseudomonas Aeruginosa Inhibits the Growth of Cryptococcus Species. Mycopathologia 2012, 173, 451–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, B.; Emami, A.; Zhou, R.; Lang, S.; Duan, Y.; Wang, Y.; Jiang, L.; Loomba, R.; Brenner, D.; Stärkel, P.; et al. Functional Microbial Responses to Alcohol Abstinence in Patients with Alcohol Use Disorder. Front. Physiol. 2020, 11. [Google Scholar] [CrossRef] [Green Version]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for Enhanced Metagenomic Taxonomic Profiling. Nat. Methods 2015, 12, 902–903. [Google Scholar] [CrossRef]
- Franzosa, E.A.; McIver, L.J.; Rahnavard, G.; Thompson, L.R.; Schirmer, M.; Weingart, G.; Lipson, K.S.; Knight, R.; Caporaso, J.G.; Segata, N.; et al. Species-Level Functional Profiling of Metagenomes and Metatranscriptomes. Nat. Methods 2018, 15, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc Database of Metabolic Pathways and Enzymes—A 2019 Update. Nucleic Acids Res. 2020, 48, D445–D453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Yang, A.-M.; Inamine, T.; Hochrath, K.; Chen, P.; Wang, L.; Llorente, C.; Bluemel, S.; Hartmann, P.; Xu, J.; Koyama, Y.; et al. Intestinal Fungi Contribute to Development of Alcoholic Liver Disease. J. Clin. Investig. 2017, 127, 2829–2841. [Google Scholar] [CrossRef] [Green Version]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions between Commensal Fungi and the C-Type Lectin Receptor Dectin-1 Influence Colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef] [Green Version]
- Duan, Y.; Llorente, C.; Lang, S.; Brandl, K.; Chu, H.; Jiang, L.; White, R.C.; Clarke, T.H.; Nguyen, K.; Torralba, M.; et al. Bacteriophage Targeting of Gut Bacterium Attenuates Alcoholic Liver Disease. Nature 2019, 575, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, Z.D.; Müller, C.L.; Miraldi, E.R.; Littman, D.R.; Blaser, M.J.; Bonneau, R.A. Sparse and Compositionally Robust Inference of Microbial Ecological Networks. PLoS Comput. Biol. 2015, 11, e1004226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meinshausen, N.; Bühlmann, P. High-Dimensional Graphs and Variable Selection with the Lasso. Ann. Stat. 2006, 34, 1436–1462. [Google Scholar] [CrossRef] [Green Version]

| Non-Alcoholic Controls | Alcohol Use Disorder | Alcoholic Hepatitis | p-Value | |
|---|---|---|---|---|
| Clinical parameter | ||||
| Total n | 9 | 41 | 81 | |
| Age (years), n = 130 | 51 (27–71) | 44 (27–67) | 51 (30–75) | 0.016 * |
| Body mass index (BMI), kg/m2, n = 118 | 23 (19–29) | 24 (18–37) | 28 (19–48) | <0.001 * |
| Male gender, n (%), n = 130 | 7 (78) | 34 (83) | 54 (67) | 0.186 |
| Laboratory parameter | ||||
| Albumin (g/dL), n = 112 | 4.5 (2.2–5.2) | 2.4 (1.3–4.1) | <0.001 | |
| Alkaline phosphatase (U/L), n = 113 | 68 (33–225) | 179 (21–1153) | <0.001 | |
| ALT (U/L), n = 121 | 37 (9–184) | 45 (15–216) | 0.092 | |
| AST (U/L), n = 121 | 36 (15–283) | 132 (41–406) | <0.001 | |
| Total bilirubin (mg/dl), n = 118 | 0.5 (0.2–1.5) | 16.3 (2.5–38.6) | <0.001 | |
| GGT (U/L), n = 77 | 43 (4–1131) | 188 (33–3632) | <0.001 | |
| Platelet counts (×109/L), n = 115 | 222 (21–434) | 123 (21–447) | <0.001 | |
| Prothrombin time, s, n = 65 | 22 (11–61) | |||
| Creatinine (mg/dL), n = 118 | 0.8 (0.5–1.3) | 0.8 (0.3–8.1) | 0.791 | |
| Sodium (mEq/L), n = 79 | 133 (118–143) | |||
| INR, n = 116 | 0.9 (0.8–1.3) | 1.8 (1.0–3.7) | <0.001 | |
| FIB-4, n = 115 | 1.3 (0.4–21.4) | 8.0 (1.4–66.3) | <0.001 | |
| FIB-4 > 3.25 (F3–F4), n (%) | 6 (15) | 71 (88) |
| Treatment at Admission | Clinical Characteristics | ||||
|---|---|---|---|---|---|
| Steroids, n (%), n = 79 | 31 (39) | Model for end-stage liver disease (MELD), n = 79 | 24 (12–46) | ||
| Pentoxifylline, n (%), n = 65 | 6 (9) | MELD > 21, n (%), n = 79 | 62 (78) | ||
| Steroids and pentoxifylline, n (%), n = 65 | 1 (2) | 30-day mortality rate, n (%), n = 76 | 8 (11) | ||
| Antibiotics, n (%), n = 79 | 18 (23) | 90-day mortality rate, n (%), n = 56 | 13 (23) | ||
| Proton pump inhibitors, n (%), n = 42 | 5 (12) | Infection at admission, n (%), n = 68 | 13 (19) | ||
| Laxatives, n = 36 | 18 (50) | ||||
| Histology | |||||
| Liver biopsy available, n (%), n = 81 | 47 (58) | Bilirubinostasis, n (%), n = 44 | 0 | 14 (32) | |
| Stage of fibrosis, n (%), n = 45 | 0 | 2 (5) | 1 | 21 (48) | |
| 1 | 0 (0) | 2 | 2 (4) | ||
| 2 | 5 (11) | 3 | 7 (16) | ||
| 3 | 6 (13) | Ballooning, n (%), n = 45 | 0 | 24 (53) | |
| 4 | 32 (71) | 1 | 21 (47) | ||
| Lobular fibrosis, n (%), n = 44 | 0 | 4 (9) | Giant mitochondria, n (%), n = 41 | 0 | 35 (85) |
| 1 | 5 (11) | 1 | 6 (15) | ||
| 2 | 2 (5) | PMN infiltration, n (%), n = 45 | 0 | 10 (22) | |
| 3 | 33 (75) | 1 | 21 (47) | ||
| Pericellular fibrosis, n (%), n = 45 | 0 | 11 (24) | 2 | 14 (31) | |
| 1 | 34 (76) | Inflammatory grade, n (%), n = 46 | 0 | 11 (24) | |
| Grade of steatosis, n (%), n = 46 | 1 | 19 (41) | 1 | 26 (56) | |
| 2 | 14 (31) | 2 | 9 (20) | ||
| 3 | 13 (28) | Mallory bodies, n (%), n = 44 | 0 | 6 (14) | |
| 1 | 38 (86) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, B.; Zhang, X.; Schnabl, B. Fungi–Bacteria Correlation in Alcoholic Hepatitis Patients. Toxins 2021, 13, 143. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13020143
Gao B, Zhang X, Schnabl B. Fungi–Bacteria Correlation in Alcoholic Hepatitis Patients. Toxins. 2021; 13(2):143. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13020143
Chicago/Turabian StyleGao, Bei, Xinlian Zhang, and Bernd Schnabl. 2021. "Fungi–Bacteria Correlation in Alcoholic Hepatitis Patients" Toxins 13, no. 2: 143. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13020143





