Identification of Decrease in TRiC Proteins as Novel Targets of Alpha-Amanitin-Derived Hepatotoxicity by Comparative Proteomic Analysis In Vitro
Abstract
:1. Introduction
2. Results
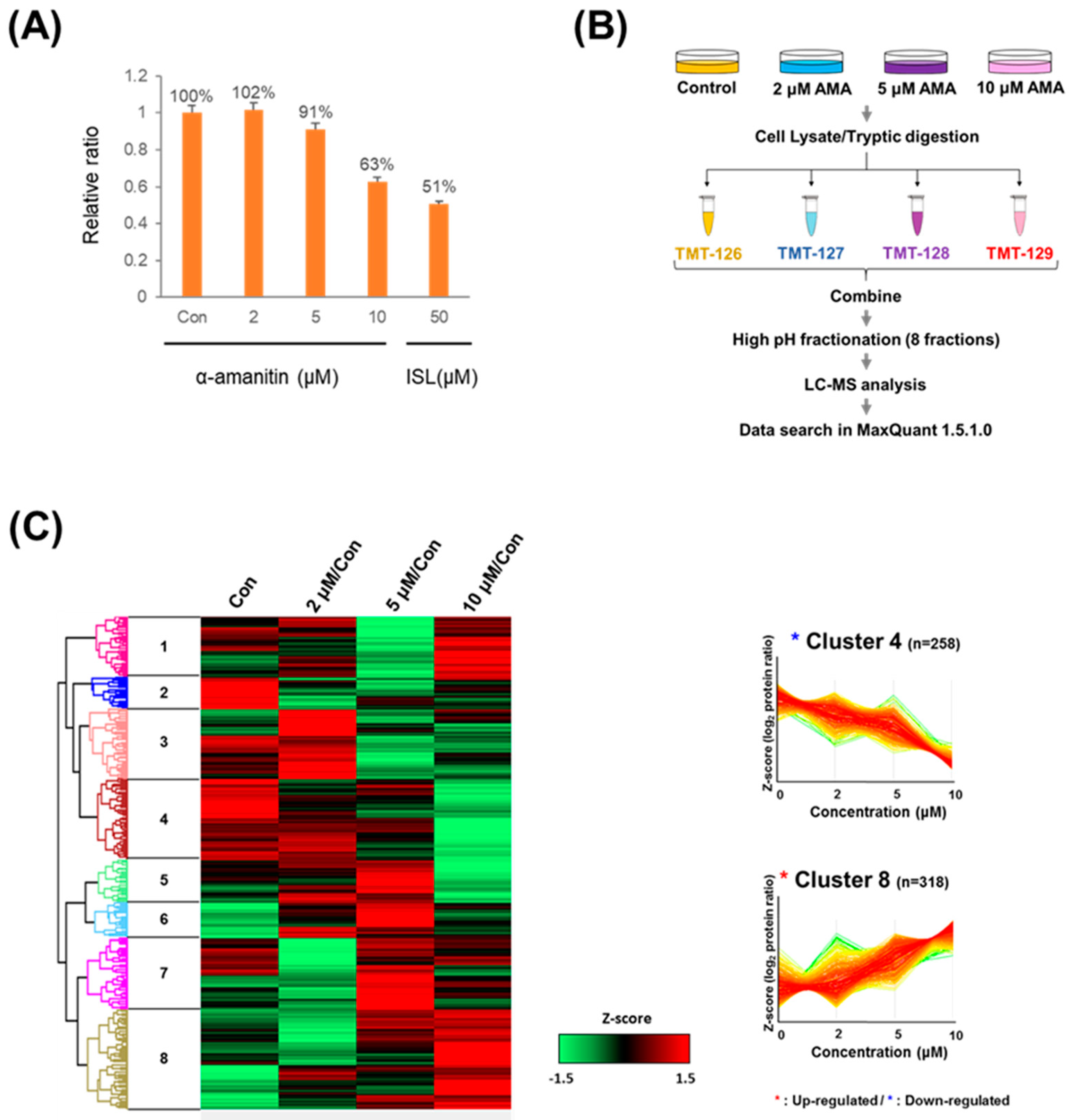
2.1. Increased Toxicity in Hepatoma Cells by α-Amanitin Treatment
2.2. Quantitative Proteomic Analysis
2.3. Identification of New Targets through Bioinformatics Analysis
2.4. Immunoblot Validation of Reduction of TRiC Proteins
3. Discussion
4. Materials and Methods
4.1. Cytotoxicity of α-AMA in Hepatoma Cells
4.2. Sample Preparation for Quantitative Proteomics Analysis
4.3. Quantitative Proteomics Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Govorushko, S.; Rezaee, R.; Dumanov, J.; Tsatsakis, A. Poisoning associated with the use of mushrooms: A review of the global pattern and main characteristics. Food Chem. Toxicol. 2019, 128, 267–279. [Google Scholar] [CrossRef]
- Abbott, N.L.; Hill, K.L.; Garrett, A.; Carter, M.D.; Hamelin, E.I.; Johnson, R.C. Detection of alpha-, beta-, and gamma-amanitin in urine by LC-MS/MS using (15)N10-alpha-amanitin as the internal standard. Toxicon 2018, 152, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Tavassoli, M.; Afshari, A.; Arsene, A.L.; Megarbane, B.; Dumanov, J.; Paoliello, M.M.B.; Tsatsakis, A.; Carvalho, F.; Hashemzaei, M.; Karimi, G.; et al. Toxicological profile of Amanita virosa—A narrative review. Toxicol. Rep. 2019, 6, 143–150. [Google Scholar] [CrossRef]
- Santi, L.; Maggioli, C.; Mastroroberto, M.; Tufoni, M.; Napoli, L.; Caraceni, P. Acute Liver Failure Caused by Amanita phalloides Poisoning. Int. J. Hepatol. 2012, 2012, 487480. [Google Scholar] [CrossRef] [PubMed]
- Letschert, K.; Faulstich, H.; Keller, D.; Keppler, D. Molecular characterization and inhibition of amanitin uptake into human hepatocytes. Toxicol. Sci. 2006, 91, 140–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wieland, T. The toxic peptides from Amanita mushrooms. Int. J. Pept. Protein Res. 1983, 22, 257–276. [Google Scholar] [CrossRef] [PubMed]
- Michelot, D.; Labia, R. Alpha-amanitin: A possible suicide substrate-like toxin involving the sulphoxide moiety of the bridged cyclopeptide. Drug Metabol. Drug Interact. 1988, 6, 265–274. [Google Scholar] [CrossRef]
- Meinecke, B.; Meinecke-Tillmann, S. Effects of alpha-amanitin on nuclear maturation of porcine oocytes in vitro. J. Reprod Fertil. 1993, 98, 195–201. [Google Scholar] [CrossRef]
- Mas, A. Mushrooms, amatoxins and the liver. J. Hepatol. 2005, 42, 166–169. [Google Scholar] [CrossRef]
- Schmutz, M.; Carron, P.N.; Yersin, B.; Trueb, L. Mushroom poisoning: A retrospective study concerning 11-years of admissions in a Swiss Emergency Department. Intern. Emerg. Med. 2018, 13, 59–67. [Google Scholar] [CrossRef]
- Moldenhauer, G.; Salnikov, A.V.; Luttgau, S.; Herr, I.; Anderl, J.; Faulstich, H. Therapeutic potential of amanitin-conjugated anti-epithelial cell adhesion molecule monoclonal antibody against pancreatic carcinoma. J. Natl. Cancer Inst. 2012, 104, 622–634. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Chen, Y.; Guo, Z.; Yang, C.; Qi, J.; Fu, Y.; Chen, Z.; Chen, P.; Wang, Y. Changes in the mitochondrial proteome in human hepatocytes in response to alpha-amanitin hepatotoxicity. Toxicon 2018, 156, 34–40. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Fujibuchi, W.; Kanehisa, M. Computation with the KEGG pathway database. Biosystems 1998, 47, 119–128. [Google Scholar] [CrossRef]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37, D211–215. [Google Scholar] [CrossRef] [Green Version]
- Gene Ontology, C. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Ditzel, L.; Lowe, J.; Stock, D.; Stetter, K.O.; Huber, H.; Huber, R.; Steinbacher, S. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell 1998, 93, 125–138. [Google Scholar] [CrossRef] [Green Version]
- Pereira, J.H.; Ralston, C.Y.; Douglas, N.R.; Meyer, D.; Knee, K.M.; Goulet, D.R.; King, J.A.; Frydman, J.; Adams, P.D. Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle. J. Biol. Chem. 2010, 285, 27958–27966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leitner, A.; Joachimiak, L.A.; Bracher, A.; Monkemeyer, L.; Walzthoeni, T.; Chen, B.; Pechmann, S.; Holmes, S.; Cong, Y.; Ma, B.; et al. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 2012, 20, 814–825. [Google Scholar] [CrossRef] [Green Version]
- Llorca, O.; Martin-Benito, J.; Ritco-Vonsovici, M.; Grantham, J.; Hynes, G.M.; Willison, K.R.; Carrascosa, J.L.; Valpuesta, J.M. Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J. 2000, 19, 5971–5979. [Google Scholar] [CrossRef] [Green Version]
- Young, J.C.; Agashe, V.R.; Siegers, K.; Hartl, F.U. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 2004, 5, 781–791. [Google Scholar] [CrossRef]
- Munoz, I.G.; Yebenes, H.; Zhou, M.; Mesa, P.; Serna, M.; Park, A.Y.; Bragado-Nilsson, E.; Beloso, A.; de Carcer, G.; Malumbres, M.; et al. Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat. Struct. Mol. Biol. 2011, 18, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Sot, B.; Rubio-Munoz, A.; Leal-Quintero, A.; Martinez-Sabando, J.; Marcilla, M.; Roodveldt, C.; Valpuesta, J.M. The chaperonin CCT inhibits assembly of alpha-synuclein amyloid fibrils by a specific, conformation-dependent interaction. Sci. Rep. 2017, 7, 40859. [Google Scholar] [CrossRef]
- Huang, X.; Wang, X.; Cheng, C.; Cai, J.; He, S.; Wang, H.; Liu, F.; Zhu, C.; Ding, Z.; Huang, X. Chaperonin containing tcp 1, subunit 8 (cct 8) is upregulated in hepatocellular carcinoma and promotes hcc proliferation. Apmis 2014, 122, 1070–1079. [Google Scholar] [CrossRef] [PubMed]
- Coghlin, C.; Carpenter, B.; Dundas, S.R.; Lawrie, L.C.; Telfer, C.; Murray, G.I. Characterization and over-expression of chaperonin t-complex proteins in colorectal cancer. J. Pathol. 2006, 210, 351–357. [Google Scholar] [CrossRef]
- Sontag, E.M.; Joachimiak, L.A.; Tan, Z.; Tomlinson, A.; Housman, D.E.; Glabe, C.G.; Potkin, S.G.; Frydman, J.; Thompson, L.M. Exogenous delivery of chaperonin subunit fragment ApiCCT1 modulates mutant Huntingtin cellular phenotypes. Proc. Natl. Acad. Sci. USA 2013, 110, 3077–3082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokota, S.; Yamamoto, Y.; Shimizu, K.; Momoi, H.; Kamikawa, T.; Yamaoka, Y.; Yanagi, H.; Yura, T.; Kubota, H. Increased expression of cytosolic chaperonin CCT in human hepatocellular and colonic carcinoma. Cell Stress Chaperones 2001, 6, 345–350. [Google Scholar] [CrossRef] [Green Version]
- Ooe, A.; Kato, K.; Noguchi, S. Possible involvement of CCT5, RGS3, and YKT6 genes up-regulated in p53-mutated tumors in resistance to docetaxel in human breast cancers. Breast Cancer Res. Treat. 2007, 101, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Khabirova, E.; Moloney, A.; Marciniak, S.J.; Williams, J.; Lomas, D.A.; Oliver, S.G.; Favrin, G.; Sattelle, D.B.; Crowther, D.C. The TRiC/CCT chaperone is implicated in Alzheimer’s disease based on patient GWAS and an RNAi screen in Aβ-expressing Caenorhabditis elegans. PLoS ONE 2014, 9, e102985. [Google Scholar] [CrossRef] [Green Version]
- Rademacher, F.; Kehren, V.; Stoldt, V.R.; Ernst, J.F. A Candida albicans chaperonin subunit (CaCct8p) as a suppressor of morphogenesis and Ras phenotypes in C-albicans and Saccharomyces cerevisiae. Microbiol. UK 1998, 144, 2951–2960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavel, M.; Imarisio, S.; Menzies, F.M.; Jimenez-Sanchez, M.; Siddiqi, F.H.; Wu, X.; Renna, M.; O’Kane, C.J.; Crowther, D.C.; Rubinsztein, D.C. CCT complex restricts neuropathogenic protein aggregation via autophagy. Nat. Commun. 2016, 7, 13821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontanella, B.; Birolo, L.; Infusini, G.; Cirulli, C.; Marzullo, L.; Pucci, P.; Turco, M.C.; Tosco, A. The co-chaperone BAG3 interacts with the cytosolic chaperonin CCT: New hints for actin folding. Int. J. Biochem. Cell Biol. 2010, 42, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Kasembeli, M.; Lau, W.C.; Roh, S.H.; Eckols, T.K.; Frydman, J.; Chiu, W.; Tweardy, D.J. Modulation of STAT3 folding and function by TRiC/CCT chaperonin. PLoS Biol. 2014, 12, e1001844. [Google Scholar] [CrossRef]
- Liu, B.D.; Larsson, L.; Caballero, A.; Hao, X.X.; Oling, D.; Grantham, J.; Nystrom, T. The Polarisome Is Required for Segregation and Retrograde Transport of Protein Aggregates. Cell 2010, 140, 257–267. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.R.; Luo, Y.H.; Piao, X.J.; Zhang, Y.; Feng, Y.C.; Li, J.Q.; Xu, W.T.; Zhang, Y.; Zhang, T.; Wang, S.N.; et al. Mechanisms underlying isoliquiritigenin-induced apoptosis and cell cycle arrest via ROS-mediated MAPK/STAT3/NF-kappaB pathways in human hepatocellular carcinoma cells. Drug Dev. Res. 2019, 80, 461–470. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.; Kim, S.; Na, A.-Y.; Sohn, C.H.; Lee, S.; Lee, H.S. Identification of Decrease in TRiC Proteins as Novel Targets of Alpha-Amanitin-Derived Hepatotoxicity by Comparative Proteomic Analysis In Vitro. Toxins 2021, 13, 197. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13030197
Kim D, Kim S, Na A-Y, Sohn CH, Lee S, Lee HS. Identification of Decrease in TRiC Proteins as Novel Targets of Alpha-Amanitin-Derived Hepatotoxicity by Comparative Proteomic Analysis In Vitro. Toxins. 2021; 13(3):197. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13030197
Chicago/Turabian StyleKim, Doeun, Sunjoo Kim, Ann-Yae Na, Chang Hwan Sohn, Sangkyu Lee, and Hye Suk Lee. 2021. "Identification of Decrease in TRiC Proteins as Novel Targets of Alpha-Amanitin-Derived Hepatotoxicity by Comparative Proteomic Analysis In Vitro" Toxins 13, no. 3: 197. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13030197





