Shiga Toxins: An Update on Host Factors and Biomedical Applications
Abstract
:1. Introduction
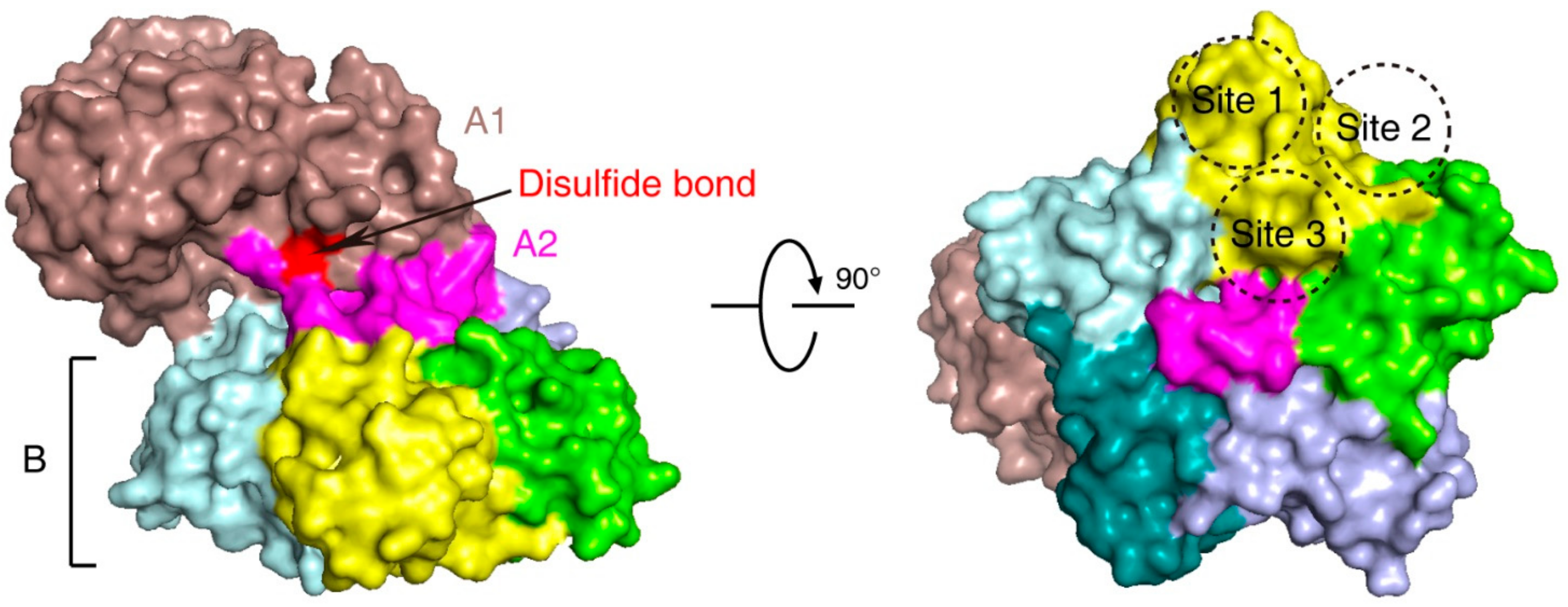
2. Stx Structure and Function
3. Stx Subtypes
4. In Vitro Cultured Human Cell Models and In Vivo Animal Models
5. Gb3 Expression in Humans and Hemolytic Uremic Syndrome (HUS)
6. Host Factors Recently Identified through CRISPR-Cas9 Screens
7. Biomedical Application of Shiga Toxin Subunit A
8. Biomedical Application of Shiga Toxin Subunit B
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Trofa, A.F.; Ueno-Olsen, H.; Oiwa, R.; Yoshikawa, M. Dr. Kiyoshi Shiga: Discoverer of the dysentery bacillus. Clin. Infect. Dis. 1999, 29, 1303–1306. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, M.B.; Estrada-Garcia, T. Shigella: A Highly Virulent and Elusive Pathogen. Curr. Trop. Med. Rep. 2014, 1, 81–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konowalchuk, J.; Speirs, J.I.; Stavric, S. Vero response to a cytotoxin of Escherichia coli. Infect. Immun. 1977, 18, 775–779. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, A.D.; Newland, J.W.; Miller, S.F.; Holmes, R.K.; Smith, H.W.; Formal, S.B. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 1984, 226, 694–696. [Google Scholar] [CrossRef]
- Jackson, M.P.; Neill, R.J.; O’Brien, A.D.; Holmes, R.K.; Newland, J.W. Nucleotide sequence analysis and comparison of the structural genes for Shiga-like toxin I and Shiga-like toxin II encoded by bacteriophages from Escherichia coli 933. FEMS Microbiol. Lett. 1987, 44, 109–114. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). National Shiga Toxin-Producing Escherichia coli (STEC) Surveillance Overview. Atlanta, Georgia: US Department of Health and Human Services, CDC. 2012. Available online: https://www.cdc.gov/nationalsurveillance/ecoli-surveillance/html (accessed on 17 March 2021).
- Tarr, P.I.; Gordon, C.A.; Chandler, W.L. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 2005, 365, 1073–1086. [Google Scholar] [CrossRef]
- Boyce, T.G.; Swerdlow, D.L.; Griffin, P.M. Escherichia coli O157:H7 and the hemolytic-uremic syndrome. N. Engl. J. Med. 1995, 333, 364–368. [Google Scholar] [CrossRef]
- Joseph, A.; Cointe, A.; Mariani Kurkdjian, P.; Rafat, C.; Hertig, A. Shiga Toxin-Associated Hemolytic Uremic Syndrome: A Narrative Review. Toxins 2020, 12, 67. [Google Scholar] [CrossRef] [Green Version]
- Williams, D.M.; Sreedhar, S.S.; Mickell, J.J.; Chan, J.C. Acute kidney failure: A pediatric experience over 20 years. Arch. Pediatr. Adolesc. Med. 2002, 156, 893–900. [Google Scholar] [CrossRef] [Green Version]
- Johannes, L.; Romer, W. Shiga toxins--from cell biology to biomedical applications. Nat. Rev. Microbiol. 2010, 8, 105–116. [Google Scholar] [CrossRef]
- Obrig, T.G. Escherichia coli Shiga Toxin Mechanisms of Action in Renal Disease. Toxins 2010, 2, 2769–2794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engedal, N.; Skotland, T.; Torgersen, M.L.; Sandvig, K. Shiga toxin and its use in targeted cancer therapy and imaging. Microb. Biotechnol. 2011, 4, 32–46. [Google Scholar] [CrossRef]
- Bergan, J.; Dyve Lingelem, A.B.; Simm, R.; Skotland, T.; Sandvig, K. Shiga toxins. Toxicon 2012, 60, 1085–1107. [Google Scholar] [CrossRef]
- Obrig, T.G.; Karpman, D. Shiga toxin pathogenesis: Kidney complications and renal failure. Curr. Top. Microbiol. Immunol. 2012, 357, 105–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacheco, A.R.; Sperandio, V. Shiga toxin in enterohemorrhagic E.coli: Regulation and novel anti-virulence strategies. Front. Cell Infect. Microbiol. 2012, 2, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melton-Celsa, A.R. Shiga Toxin (Stx) Classification, Structure, and Function. Microbiol. Spectr. 2014, 2, 37–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, Y.J.; Park, S.K.; Yoon, S.J.; Park, Y.J.; Lee, M.S. Experimental In Vivo Models of Bacterial Shiga Toxin-Associated Hemolytic Uremic Syndrome. J. Microbiol. Biotechnol. 2018, 28, 1413–1425. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Tesh, V.L. Roles of Shiga Toxins in Immunopathology. Toxins 2019, 11, 212. [Google Scholar] [CrossRef] [Green Version]
- Lingwood, C. Verotoxin Receptor-Based Pathology and Therapies. Front. Cell Infect. Microbiol. 2020, 10, 123. [Google Scholar] [CrossRef] [Green Version]
- Luginbuehl, V.; Meier, N.; Kovar, K.; Rohrer, J. Intracellular drug delivery: Potential usefulness of engineered Shiga toxin subunit B for targeted cancer therapy. Biotechnol. Adv. 2018, 36, 613–623. [Google Scholar] [CrossRef]
- Menge, C. Molecular Biology of Escherichia Coli Shiga Toxins’ Effects on Mammalian Cells. Toxins 2020, 12, 345. [Google Scholar] [CrossRef]
- Fraser, M.E.; Chernaia, M.M.; Kozlov, Y.V.; James, M.N. Crystal structure of the holotoxin from Shigella dysenteriae at 2.5 A resolution. Nat. Struct. Biol. 1994, 1, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Stein, P.E.; Boodhoo, A.; Tyrrell, G.J.; Brunton, J.L.; Read, R.J. Crystal structure of the cell-binding B oligomer of verotoxin-1 from E. coli. Nature 1992, 355, 748–750. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Tsurugi, K.; Yutsudo, T.; Takeda, Y.; Ogasawara, T.; Igarashi, K. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur. J. Biochem. 1988, 171, 45–50. [Google Scholar] [CrossRef]
- Saxena, S.K.; O’Brien, A.D.; Ackerman, E.J. Shiga toxin, Shiga-like toxin II variant, and ricin are all single-site RNA N-glycosidases of 28 S RNA when microinjected into Xenopus oocytes. J. Biol. Chem. 1989, 264, 596–601. [Google Scholar] [CrossRef]
- Ogasawara, T.; Ito, K.; Igarashi, K.; Yutsudo, T.; Nakabayashi, N.; Takeda, Y. Inhibition of protein synthesis by a Vero toxin (VT2 or Shiga-like toxin II) produced by Escherichia coli O157:H7 at the level of elongation factor 1-dependent aminoacyl-tRNA binding to ribosomes. Microb. Pathog. 1988, 4, 127–135. [Google Scholar] [CrossRef]
- Lindberg, A.A.; Brown, J.E.; Stromberg, N.; Westling-Ryd, M.; Schultz, J.E.; Karlsson, K.A. Identification of the carbohydrate receptor for Shiga toxin produced by Shigella dysenteriae type 1. J. Biol. Chem. 1987, 262, 1779–1785. [Google Scholar] [CrossRef]
- Ling, H.; Boodhoo, A.; Hazes, B.; Cummings, M.D.; Armstrong, G.D.; Brunton, J.L.; Read, R.J. Structure of the shiga-like toxin I B-pentamer complexed with an analogue of its receptor Gb3. Biochemistry 1998, 37, 1777–1788. [Google Scholar] [CrossRef]
- Romer, W.; Pontani, L.L.; Sorre, B.; Rentero, C.; Berland, L.; Chambon, V.; Lamaze, C.; Bassereau, P.; Sykes, C.; Gaus, K.; et al. Actin dynamics drive membrane reorganization and scission in clathrin-independent endocytosis. Cell 2010, 140, 540–553. [Google Scholar] [CrossRef] [Green Version]
- Sandvig, K.; van Deurs, B. Endocytosis, intracellular transport, and cytotoxic action of Shiga toxin and ricin. Physiol. Rev. 1996, 76, 949–966. [Google Scholar] [CrossRef]
- Sandvig, K.; Olsnes, S.; Brown, J.E.; Petersen, O.W.; van Deurs, B. Endocytosis from coated pits of Shiga toxin: A glycolipid-binding protein from Shigella dysenteriae 1. J. Cell Biol. 1989, 108, 1331–1343. [Google Scholar] [CrossRef] [Green Version]
- Saint-Pol, A.; Yelamos, B.; Amessou, M.; Mills, I.G.; Dugast, M.; Tenza, D.; Schu, P.; Antony, C.; McMahon, H.T.; Lamaze, C.; et al. Clathrin adaptor epsinR is required for retrograde sorting on early endosomal membranes. Dev. Cell 2004, 6, 525–538. [Google Scholar] [CrossRef] [Green Version]
- Johannes, L.; Popoff, V. Tracing the retrograde route in protein trafficking. Cell 2008, 135, 1175–1187. [Google Scholar] [CrossRef] [Green Version]
- Sandvig, K.; Skotland, T.; van Deurs, B.; Klokk, T.I. Retrograde transport of protein toxins through the Golgi apparatus. Histochem. Cell Biol. 2013, 140, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Mallard, F.; Antony, C.; Tenza, D.; Salamero, J.; Goud, B.; Johannes, L. Direct pathway from early/recycling endosomes to the Golgi apparatus revealed through the study of shiga toxin B-fragment transport. J. Cell Biol. 1998, 143, 973–990. [Google Scholar] [CrossRef]
- Garred, O.; Dubinina, E.; Polesskaya, A.; Olsnes, S.; Kozlov, J.; Sandvig, K. Role of the disulfide bond in Shiga toxin A-chain for toxin entry into cells. J. Biol. Chem. 1997, 272, 11414–11419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fagerquist, C.K.; Sultan, O. Top-down proteomic identification of furin-cleaved alpha-subunit of Shiga toxin 2 from Escherichia coli O157:H7 using MALDI-TOF-TOF-MS/MS. J. Biomed. Biotechnol. 2010, 2010, 123460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheutz, F.; Teel, L.D.; Beutin, L.; Pierard, D.; Buvens, G.; Karch, H.; Mellmann, A.; Caprioli, A.; Tozzoli, R.; Morabito, S.; et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 2012, 50, 2951–2963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, X.; Fu, S.; Zhang, J.; Fan, R.; Xu, Y.; Sun, H.; He, X.; Xu, J.; Xiong, Y. Identification and pathogenomic analysis of an Escherichia coli strain producing a novel Shiga toxin 2 subtype. Sci. Rep. 2018, 8, 6756. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.C.; Zhang, Y.; Bai, X.; Xiong, Y.; Wang, Y.; Yang, X.; Xu, Q.; He, X. Structural and Functional Characterization of Stx2k, a New Subtype of Shiga Toxin 2. Microorganisms 2019, 8, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Bai, X.; Zhang, J.; Sun, H.; Fu, S.; Fan, R.; He, X.; Scheutz, F.; Matussek, A.; Xiong, Y. Escherichia coli strains producing a novel Shiga toxin 2 subtype circulate in China. Int. J. Med. Microbiol. 2020, 310, 151377. [Google Scholar] [CrossRef] [PubMed]
- Fuller, C.A.; Pellino, C.A.; Flagler, M.J.; Strasser, J.E.; Weiss, A.A. Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 2011, 79, 1329–1337. [Google Scholar] [CrossRef] [Green Version]
- Luna-Gierke, R.E.; Griffin, P.M.; Gould, L.H.; Herman, K.; Bopp, C.A.; Strockbine, N.; Mody, R.K. Outbreaks of non-O157 Shiga toxin-producing Escherichia coli infection: USA. Epidemiol. Infect. 2014, 142, 2270–2280. [Google Scholar] [CrossRef]
- Fraser, M.E.; Fujinaga, M.; Cherney, M.M.; Melton-Celsa, A.R.; Twiddy, E.M.; O’Brien, A.D.; James, M.N. Structure of shiga toxin type 2 (Stx2) from Escherichia coli O157:H7. J. Biol. Chem. 2004, 279, 27511–27517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casanova, N.A.; Redondo, L.M.; Dailoff, G.C.; Arenas, D.; Fernandez Miyakawa, M.E. Overview of the role of Shiga toxins in porcine edema disease pathogenesis. Toxicon 2018, 148, 149–154. [Google Scholar] [CrossRef]
- Farooq, S.; Hussain, I.; Mir, M.A.; Bhat, M.A.; Wani, S.A. Isolation of atypical enteropathogenic Escherichia coli and Shiga toxin 1 and 2f-producing Escherichia coli from avian species in India. Lett. Appl. Microbiol. 2009, 48, 692–697. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.; Etoh, Y.; Ichihara, S.; Maeda, E.; Takenaka, S.; Horikawa, K.; Narimatsu, H.; Kawano, K.; Kawamura, Y.; Ito, K. Isolation and characteristics of Shiga toxin 2f-producing Escherichia coli among pigeons in Kyushu, Japan. PLoS ONE 2014, 9, e86076. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, H.; Scheef, J.; Morabito, S.; Caprioli, A.; Wieler, L.H.; Karch, H. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 2000, 66, 1205–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacher, D.W.; Gangiredla, J.; Patel, I.; Elkins, C.A.; Feng, P.C. Use of the Escherichia coli Identification Microarray for Characterizing the Health Risks of Shiga Toxin-Producing Escherichia coli Isolated from Foods. J. Food Prot. 2016, 79, 1656–1662. [Google Scholar] [CrossRef] [PubMed]
- Tyrrell, G.J.; Ramotar, K.; Toye, B.; Boyd, B.; Lingwood, C.A.; Brunton, J.L. Alteration of the carbohydrate binding specificity of verotoxins from Gal alpha 1-4Gal to GalNAc beta 1-3Gal alpha 1-4Gal and vice versa by site-directed mutagenesis of the binding subunit. Proc. Natl. Acad. Sci. USA 1992, 89, 524–528. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, H.; Kiyokawa, N.; Katagiri, Y.U.; Taguchi, T.; Suzuki, T.; Sekino, T.; Mimori, K.; Ebata, T.; Saito, M.; Nakao, H.; et al. Kinetic analysis of binding between Shiga toxin and receptor glycolipid Gb3Cer by surface plasmon resonance. J. Biol. Chem. 2001, 276, 42915–42922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedrich, A.W.; Bielaszewska, M.; Zhang, W.L.; Pulz, M.; Kuczius, T.; Ammon, A.; Karch, H. Escherichia coli harboring Shiga toxin 2 gene variants: Frequency and association with clinical symptoms. J. Infect. Dis. 2002, 185, 74–84. [Google Scholar] [CrossRef] [Green Version]
- Viel, T.; Dransart, E.; Nemati, F.; Henry, E.; Thézé, B.; Decaudin, D.; Lewandowski, D.; Boisgard, R.; Johannes, L.; Tavitian, B. In Vivo Tumor Targeting by the B-Subunit of Shiga Toxin. Mol. Imaging 2008, 7, 239–247. [Google Scholar] [CrossRef]
- Kouzel, I.U.; Pohlentz, G.; Schmitz, J.S.; Steil, D.; Humpf, H.U.; Karch, H.; Muthing, J. Shiga Toxin Glycosphingolipid Receptors in Human Caco-2 and HCT-8 Colon Epithelial Cell Lines. Toxins 2017, 9, 338. [Google Scholar] [CrossRef]
- Amaral, M.M.; Sacerdoti, F.; Jancic, C.; Repetto, H.A.; Paton, A.W.; Paton, J.C.; Ibarra, C. Action of shiga toxin type-2 and subtilase cytotoxin on human microvascular endothelial cells. PLoS ONE 2013, 8, e70431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehrlenbach, S.; Rosales, A.; Posch, W.; Wilflingseder, D.; Hermann, M.; Brockmeyer, J.; Karch, H.; Satchell, S.C.; Wurzner, R.; Orth-Holler, D. Shiga toxin 2 reduces complement inhibitor CD59 expression on human renal tubular epithelial and glomerular endothelial cells. Infect. Immun. 2013, 81, 2678–2685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matussek, A.; Lauber, J.; Bergau, A.; Hansen, W.; Rohde, M.; Dittmar, K.E.; Gunzer, M.; Mengel, M.; Gatzlaff, P.; Hartmann, M.; et al. Molecular and functional analysis of Shiga toxin-induced response patterns in human vascular endothelial cells. Blood 2003, 102, 1323–1332. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Linstedt, A.D. Manganese blocks intracellular trafficking of Shiga toxin and protects against Shiga toxicosis. Science 2012, 335, 332–335. [Google Scholar] [CrossRef] [Green Version]
- Taga, S.; Carlier, K.; Mishal, Z.; Capoulade, C.; Mangeney, M.; Lecluse, Y.; Coulaud, D.; Tetaud, C.; Pritchard, L.L.; Tursz, T.; et al. Intracellular signaling events in CD77-mediated apoptosis of Burkitt’s lymphoma cells. Blood 1997, 90, 2757–2767. [Google Scholar] [CrossRef]
- Brandelli, J.R.; Griener, T.P.; Laing, A.; Mulvey, G.; Armstrong, G.D. The Effects of Shiga Toxin 1, 2 and Their Subunits on Cytokine and Chemokine Expression by Human Macrophage-Like THP-1 Cells. Toxins 2015, 7, 4054–4066. [Google Scholar] [CrossRef] [Green Version]
- Johansson, K.; Willysson, A.; Kristoffersson, A.C.; Tontanahal, A.; Gillet, D.; Stahl, A.L.; Karpman, D. Shiga Toxin-Bearing Microvesicles Exert a Cytotoxic Effect on Recipient Cells Only When the Cells Express the Toxin Receptor. Front. Cell Infect. Microbiol. 2020, 10, 212. [Google Scholar] [CrossRef]
- Waddell, T.; Cohen, A.; Lingwood, C.A. Induction of verotoxin sensitivity in receptor-deficient cell lines using the receptor glycolipid globotriosylceramide. Proc. Natl. Acad. Sci. USA 1990, 87, 7898–7901. [Google Scholar] [CrossRef] [Green Version]
- Tian, S.; Muneeruddin, K.; Choi, M.Y.; Tao, L.; Bhuiyan, R.H.; Ohmi, Y.; Furukawa, K.; Furukawa, K.; Boland, S.; Shaffer, S.A.; et al. Genome-wide CRISPR screens for Shiga toxins and ricin reveal Golgi proteins critical for glycosylation. PLoS Biol. 2018, 16, e2006951. [Google Scholar] [CrossRef] [Green Version]
- Lingwood, C.A.; Binnington, B.; Manis, A.; Branch, D.R. Globotriaosyl ceramide receptor function—Where membrane structure and pathology intersect. FEBS Lett. 2010, 584, 1879–1886. [Google Scholar] [CrossRef] [Green Version]
- Lingwood, C.A.; Manis, A.; Mahfoud, R.; Khan, F.; Binnington, B.; Mylvaganam, M. New aspects of the regulation of glycosphingolipid receptor function. Chem. Phys. Lipids 2010, 163, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Obrig, T.G.; Louise, C.B.; Lingwood, C.A.; Boyd, B.; Barley-Maloney, L.; Daniel, T.O. Endothelial heterogeneity in Shiga toxin receptors and responses. J. Biol. Chem. 1993, 268, 15484–15488. [Google Scholar] [CrossRef]
- Kouzel, I.U.; Kehl, A.; Berger, P.; Liashkovich, I.; Steil, D.; Makalowski, W.; Suzuki, Y.; Pohlentz, G.; Karch, H.; Mellmann, A.; et al. RAB5A and TRAPPC6B are novel targets for Shiga toxin 2a inactivation in kidney epithelial cells. Sci. Rep. 2020, 10, 4945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, C.L.; Mahe, M.M.; Munera, J.; Howell, J.C.; Sundaram, N.; Poling, H.M.; Schweitzer, J.I.; Vallance, J.E.; Mayhew, C.N.; Sun, Y.; et al. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 2014, 20, 1310–1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karve, S.S.; Pradhan, S.; Ward, D.V.; Weiss, A.A. Intestinal organoids model human responses to infection by commensal and Shiga toxin producing Escherichia coli. PLoS ONE 2017, 12, e0178966. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, S.; Karve, S.S.; Weiss, A.A.; Hawkins, J.; Poling, H.M.; Helmrath, M.A.; Wells, J.M.; McCauley, H.A. Tissue Responses to Shiga Toxin in Human Intestinal Organoids. Cell Mol. Gastroenterol. Hepatol. 2020, 10, 171–190. [Google Scholar] [CrossRef] [Green Version]
- Mohawk, K.L.; O’Brien, A.D. Mouse models of Escherichia coli O157:H7 infection and shiga toxin injection. J. Biomed. Biotechnol. 2011, 2011, 258185. [Google Scholar] [CrossRef] [Green Version]
- Okuda, T.; Tokuda, N.; Numata, S.; Ito, M.; Ohta, M.; Kawamura, K.; Wiels, J.; Urano, T.; Tajima, O.; Furukawa, K.; et al. Targeted disruption of Gb3/CD77 synthase gene resulted in the complete deletion of globo-series glycosphingolipids and loss of sensitivity to verotoxins. J. Biol. Chem. 2006, 281, 10230–10235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paixao-Cavalcante, D.; Botto, M.; Cook, H.T.; Pickering, M.C. Shiga toxin-2 results in renal tubular injury but not thrombotic microangiopathy in heterozygous factor H-deficient mice. Clin. Exp. Immunol. 2009, 155, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Keepers, T.R.; Psotka, M.A.; Gross, L.K.; Obrig, T.G. A murine model of HUS: Shiga toxin with lipopolysaccharide mimics the renal damage and physiologic response of human disease. J. Am. Soc. Nephrol. 2006, 17, 3404–3414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.S.; Kwon, H.; Lee, E.Y.; Kim, D.J.; Park, J.H.; Tesh, V.L.; Oh, T.K.; Kim, M.H. Shiga Toxins Activate the NLRP3 Inflammasome Pathway To Promote Both Production of the Proinflammatory Cytokine Interleukin-1beta and Apoptotic Cell Death. Infect. Immun. 2016, 84, 172–186. [Google Scholar] [CrossRef] [Green Version]
- Havira, M.S.; Ta, A.; Kumari, P.; Wang, C.; Russo, A.J.; Ruan, J.; Rathinam, V.A.; Vanaja, S.K. Shiga toxin suppresses noncanonical inflammasome responses to cytosolic LPS. Sci. Immunol. 2020, 5, eabc0217. [Google Scholar] [CrossRef]
- Wadolkowski, E.A.; Burris, J.A.; O’Brien, A.D. Mouse model for colonization and disease caused by enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 1990, 58, 2438–2445. [Google Scholar] [CrossRef] [Green Version]
- Lindgren, S.W.; Melton, A.R.; O’Brien, A.D. Virulence of enterohemorrhagic Escherichia coli O91:H21 clinical isolates in an orally infected mouse model. Infect. Immun. 1993, 61, 3832–3842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, K.; Asahara, T.; Nomoto, K.; Tanaka, R.; Hamabata, T.; Ozawa, A.; Takeda, Y. Development of a lethal Shiga toxin-producing Escherichia coli-infection mouse model using multiple mitomycin C treatment. Microb. Pathog. 2003, 35, 1–9. [Google Scholar] [CrossRef]
- Eaton, K.A.; Friedman, D.I.; Francis, G.J.; Tyler, J.S.; Young, V.B.; Haeger, J.; Abu-Ali, G.; Whittam, T.S. Pathogenesis of renal disease due to enterohemorrhagic Escherichia coli in germ-free mice. Infect. Immun. 2008, 76, 3054–3063. [Google Scholar] [CrossRef] [Green Version]
- Mallick, E.M.; McBee, M.E.; Vanguri, V.K.; Melton-Celsa, A.R.; Schlieper, K.; Karalius, B.J.; O’Brien, A.D.; Butterton, J.R.; Leong, J.M.; Schauer, D.B. A novel murine infection model for Shiga toxin-producing Escherichia coli. J. Clin. Investig. 2012, 122, 4012–4024. [Google Scholar] [CrossRef]
- Sugatani, J.; Komiyama, N.; Mochizuki, T.; Hoshino, M.; Miyamoto, D.; Igarashi, T.; Hoshi, S.; Miwa, M. Urinary concentrating defect in rats given Shiga toxin: Elevation in urinary AQP2 level associated with polyuria. Life Sci. 2002, 71, 171–189. [Google Scholar] [CrossRef]
- Zotta, E.; Lago, N.; Ochoa, F.; Repetto, H.A.; Ibarra, C. Development of an experimental hemolytic uremic syndrome in rats. Pediatr. Nephrol. 2008, 23, 559–567. [Google Scholar] [CrossRef]
- Silberstein, C.; Lucero, M.S.; Zotta, E.; Copeland, D.P.; Lingyun, L.; Repetto, H.A.; Ibarra, C. A glucosylceramide synthase inhibitor protects rats against the cytotoxic effects of shiga toxin 2. Pediatr. Res. 2011, 69, 390–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stone, S.M.; Thorpe, C.M.; Ahluwalia, A.; Rogers, A.B.; Obata, F.; Vozenilek, A.; Kolling, G.L.; Kane, A.V.; Magun, B.E.; Jandhyala, D.M. Shiga toxin 2-induced intestinal pathology in infant rabbits is A-subunit dependent and responsive to the tyrosine kinase and potential ZAK inhibitor imatinib. Front. Cell Infect. Microbiol. 2012, 2, 135. [Google Scholar] [CrossRef] [Green Version]
- Garcia, A.; Bosques, C.J.; Wishnok, J.S.; Feng, Y.; Karalius, B.J.; Butterton, J.R.; Schauer, D.B.; Rogers, A.B.; Fox, J.G. Renal injury is a consistent finding in Dutch Belted rabbits experimentally infected with enterohemorrhagic Escherichia coli. J. Infect. Dis. 2006, 193, 1125–1134. [Google Scholar] [CrossRef] [Green Version]
- Garcia, A.; Marini, R.P.; Catalfamo, J.L.; Knox, K.A.; Schauer, D.B.; Rogers, A.B.; Fox, J.G. Intravenous Shiga toxin 2 promotes enteritis and renal injury characterized by polymorphonuclear leukocyte infiltration and thrombosis in Dutch Belted rabbits. Microbes Infect. 2008, 10, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.; Pulimood, A.B.; Koshi, R.; Hull, A.; Acheson, D.; Rajan, P.; Keusch, G.T.; Mathan, V.I.; Mathan, M.M. A monkey model for enterohemorrhagic Escherichia coli infection. J. Infect. Dis. 2001, 184, 206–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegler, R.L.; Pysher, T.J.; Tesh, V.L.; Taylor, F.B., Jr. Response to single and divided doses of Shiga toxin-1 in a primate model of hemolytic uremic syndrome. J. Am. Soc. Nephrol. 2001, 12, 1458–1467. [Google Scholar]
- Siegler, R.L.; Obrig, T.G.; Pysher, T.J.; Tesh, V.L.; Denkers, N.D.; Taylor, F.B. Response to Shiga toxin 1 and 2 in a baboon model of hemolytic uremic syndrome. Pediatr. Nephrol. 2003, 18, 92–96. [Google Scholar] [CrossRef]
- Stearns-Kurosawa, D.J.; Oh, S.Y.; Cherla, R.P.; Lee, M.S.; Tesh, V.L.; Papin, J.; Henderson, J.; Kurosawa, S. Distinct renal pathology and a chemotactic phenotype after enterohemorrhagic Escherichia coli shiga toxins in non-human primate models of hemolytic uremic syndrome. Am. J. Pathol. 2013, 182, 1227–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, F.B., Jr.; Tesh, V.L.; DeBault, L.; Li, A.; Chang, A.C.; Kosanke, S.D.; Pysher, T.J.; Siegler, R.L. Characterization of the baboon responses to Shiga-like toxin: Descriptive study of a new primate model of toxic responses to Stx-1. Am. J. Pathol. 1999, 154, 1285–1299. [Google Scholar] [CrossRef]
- Hellberg, A.; Westman, J.S.; Thuresson, B.; Olsson, M.L. P1PK: The blood group system that changed its name and expanded. Immunohematology 2013, 29, 25–33. [Google Scholar] [PubMed]
- Kasai, K.; Galton, J.; Terasaki, P.I.; Wakisaka, A.; Kawahara, M.; Root, T.; Hakomori, S.I. Tissue distribution of the Pk antigen as determined by a monoclonal antibody. J. Immunogenet 1985, 12, 213–220. [Google Scholar] [CrossRef]
- Ren, J.; Utsunomiya, I.; Taguchi, K.; Ariga, T.; Tai, T.; Ihara, Y.; Miyatake, T. Localization of verotoxin receptors in nervous system. Brain Res. 1999, 825, 183–188. [Google Scholar] [CrossRef]
- Obata, F.; Tohyama, K.; Bonev, A.D.; Kolling, G.L.; Keepers, T.R.; Gross, L.K.; Nelson, M.T.; Sato, S.; Obrig, T.G. Shiga toxin 2 affects the central nervous system through receptor globotriaosylceramide localized to neurons. J. Infect. Dis. 2008, 198, 1398–1406. [Google Scholar] [CrossRef] [Green Version]
- Wiels, J.; Fellous, M.; Tursz, T. Monoclonal antibody against a Burkitt lymphoma-associated antigen. Proc. Natl. Acad. Sci. USA 1981, 78, 6485–6488. [Google Scholar] [CrossRef] [Green Version]
- Arvidsson, I.; Stahl, A.L.; Hedstrom, M.M.; Kristoffersson, A.C.; Rylander, C.; Westman, J.S.; Storry, J.R.; Olsson, M.L.; Karpman, D. Shiga toxin-induced complement-mediated hemolysis and release of complement-coated red blood cell-derived microvesicles in hemolytic uremic syndrome. J. Immunol. 2015, 194, 2309–2318. [Google Scholar] [CrossRef] [Green Version]
- Stahl, A.L.; Arvidsson, I.; Johansson, K.E.; Chromek, M.; Rebetz, J.; Loos, S.; Kristoffersson, A.C.; Bekassy, Z.D.; Morgelin, M.; Karpman, D. A novel mechanism of bacterial toxin transfer within host blood cell-derived microvesicles. PLoS Pathog. 2015, 11, e1004619. [Google Scholar] [CrossRef] [Green Version]
- Noris, M.; Remuzzi, G. Hemolytic uremic syndrome. J. Am. Soc. Nephrol. 2005, 16, 1035–1050. [Google Scholar] [CrossRef] [Green Version]
- Noris, M.; Remuzzi, G. Atypical hemolytic-uremic syndrome. N. Engl. J. Med. 2009, 361, 1676–1687. [Google Scholar] [CrossRef]
- Keir, L.S.; Marks, S.D.; Kim, J.J. Shigatoxin-associated hemolytic uremic syndrome: Current molecular mechanisms and future therapies. Drug Des. Devel. Ther. 2012, 6, 195–208. [Google Scholar] [CrossRef] [Green Version]
- Harkins, V.J.; McAllister, D.A.; Reynolds, B.C. Shiga-Toxin, E. coli Hemolytic Uremic Syndrome: Review of Management and Long-term Outcome. Curr. Pediatr. Rep. 2020, 8, 16–25. [Google Scholar] [CrossRef] [Green Version]
- Mele, C.; Remuzzi, G.; Noris, M. Hemolytic uremic syndrome. Semin. Immunopathol. 2014, 36, 399–420. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, K.; Yokoyama, K.; Sato, T.; Wiels, J.; Hirayama, Y.; Ohta, M.; Furukawa, K. Expression of the Gb3/CD77 synthase gene in megakaryoblastic leukemia cells: Implication in the sensitivity to verotoxins. J. Biol. Chem. 2002, 277, 11247–11254. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.A.; Polanowska-Grabowska, R.K.; Fujii, J.; Obrig, T.; Gear, A.R. Shiga toxin binds to activated platelets. J. Thromb. Haemost 2004, 2, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Heyderman, R.S.; Soriani, M.; Hirst, T.R. Is immune cell activation the missing link in the pathogenesis of post-diarrhoeal HUS? Trends Microbiol. 2001, 9, 262–266. [Google Scholar] [CrossRef]
- Beneke, J.; Sartison, A.; Kielstein, J.T.; Haller, H.; Nitschke, M.; Kunzendorf, U.; Loos, S.; Kemper, M.J.; Stahl, R.A.; Menne, J.; et al. Clinical and Laboratory Consequences of Platelet Transfusion in Shiga Toxin-Mediated Hemolytic Uremic Syndrome. Transfus. Med. Rev. 2017, 31, 51–55. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelson, T.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Wei, J.J.; Sabatini, D.M.; Lander, E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 2014, 343, 80–84. [Google Scholar] [CrossRef] [Green Version]
- Pacheco, A.R.; Lazarus, J.E.; Sit, B.; Schmieder, S.; Lencer, W.I.; Blondel, C.J.; Doench, J.G.; Davis, B.M.; Waldor, M.K. CRISPR Screen Reveals that EHEC’s T3SS and Shiga Toxin Rely on Shared Host Factors for Infection. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaji, T.; Sekizuka, T.; Tachida, Y.; Sakuma, C.; Morimoto, K.; Kuroda, M.; Hanada, K. A CRISPR Screen Identifies LAPTM4A and TM9SF Proteins as Glycolipid-Regulating Factors. iScience 2019, 11, 409–424. [Google Scholar] [CrossRef] [Green Version]
- Majumder, S.; Kono, M.; Lee, Y.T.; Byrnes, C.; Li, C.; Tuymetova, G.; Proia, R.L. A genome-wide CRISPR/Cas9 screen reveals that the aryl hydrocarbon receptor stimulates sphingolipid levels. J. Biol. Chem. 2020, 295, 4341–4349. [Google Scholar] [CrossRef] [Green Version]
- Kojima, Y.; Fukumoto, S.; Furukawa, K.; Okajima, T.; Wiels, J.; Yokoyama, K.; Suzuki, Y.; Urano, T.; Ohta, M.; Furukawa, K. Molecular cloning of globotriaosylceramide/CD77 synthase, a glycosyltransferase that initiates the synthesis of globo series glycosphingolipids. J. Biol. Chem. 2000, 275, 15152–15156. [Google Scholar] [CrossRef] [Green Version]
- Keusch, J.J.; Manzella, S.M.; Nyame, K.A.; Cummings, R.D.; Baenziger, J.U. Cloning of Gb3 synthase, the key enzyme in globo-series glycosphingolipid synthesis, predicts a family of alpha 1, 4-glycosyltransferases conserved in plants, insects, and mammals. J. Biol. Chem. 2000, 275, 25315–25321. [Google Scholar] [CrossRef] [Green Version]
- Yamaji, T.; Hanada, K. Sphingolipid metabolism and interorganellar transport: Localization of sphingolipid enzymes and lipid transfer proteins. Traffic 2015, 16, 101–122. [Google Scholar] [CrossRef] [PubMed]
- Hogue, D.L.; Ellison, M.J.; Young, J.D.; Cass, C.E. Identification of a novel membrane transporter associated with intracellular membranes by phenotypic complementation in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 9801–9808. [Google Scholar] [CrossRef] [Green Version]
- Cabrita, M.A.; Hobman, T.C.; Hogue, D.L.; King, K.M.; Cass, C.E. Mouse transporter protein, a membrane protein that regulates cellular multidrug resistance, is localized to lysosomes. Cancer Res. 1999, 59, 4890–4897. [Google Scholar] [PubMed]
- Hogue, D.L.; Nash, C.; Ling, V.; Hobman, T.C. Lysosome-associated protein transmembrane 4 alpha (LAPTM4 alpha) requires two tandemly arranged tyrosine-based signals for sorting to lysosomes. Biochem. J. 2002, 365, 721–730. [Google Scholar] [CrossRef] [Green Version]
- Milkereit, R.; Rotin, D. A role for the ubiquitin ligase Nedd4 in membrane sorting of LAPTM4 proteins. PLoS ONE 2011, 6, e27478. [Google Scholar] [CrossRef] [PubMed]
- Jae, L.T.; Raaben, M.; Riemersma, M.; van Beusekom, E.; Blomen, V.A.; Velds, A.; Kerkhoven, R.M.; Carette, J.E.; Topaloglu, H.; Meinecke, P.; et al. Deciphering the glycosylome of dystroglycanopathies using haploid screens for lassa virus entry. Science 2013, 340, 479–483. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, A.; Tumkosit, U.; Nakamura, S.; Motooka, D.; Kishishita, N.; Priengprom, T.; Sa-Ngasang, A.; Kinoshita, T.; Takeda, N.; Maeda, Y. Genome-Wide Screening Uncovers the Significance of N-Sulfation of Heparan Sulfate as a Host Cell Factor for Chikungunya Virus Infection. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, L.; Tian, S.; Zhang, J.; Liu, Z.; Robinson-McCarthy, L.; Miyashita, S.I.; Breault, D.T.; Gerhard, R.; Oottamasathien, S.; Whelan, S.P.J.; et al. Sulfated glycosaminoglycans and low-density lipoprotein receptor contribute to Clostridium difficile toxin A entry into cells. Nat. Microbiol. 2019, 4, 1760–1769. [Google Scholar] [CrossRef]
- Meisen, W.H.; Nejad, Z.B.; Hardy, M.; Zhao, H.; Oliverio, O.; Wang, S.; Hale, C.; Ollmann, M.M.; Collins, P.J. Pooled Screens Identify GPR108 and TM9SF2 as Host Cell Factors Critical for AAV Transduction. Mol. Ther. Methods Clin. Dev. 2020, 17, 601–611. [Google Scholar] [CrossRef]
- Luteijn, R.D.; van Diemen, F.; Blomen, V.A.; Boer, I.G.J.; Manikam Sadasivam, S.; van Kuppevelt, T.H.; Drexler, I.; Brummelkamp, T.R.; Lebbink, R.J.; Wiertz, E.J. A Genome-Wide Haploid Genetic Screen Identifies Heparan Sulfate-Associated Genes and the Macropinocytosis Modulator TMED10 as Factors Supporting Vaccinia Virus Infection. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foulquier, F.; Amyere, M.; Jaeken, J.; Zeevaert, R.; Schollen, E.; Race, V.; Bammens, R.; Morelle, W.; Rosnoblet, C.; Legrand, D.; et al. TMEM165 deficiency causes a congenital disorder of glycosylation. Am. J. Hum. Genet. 2012, 91, 15–26. [Google Scholar] [CrossRef] [Green Version]
- Potelle, S.; Morelle, W.; Dulary, E.; Duvet, S.; Vicogne, D.; Spriet, C.; Krzewinski-Recchi, M.A.; Morsomme, P.; Jaeken, J.; Matthijs, G.; et al. Glycosylation abnormalities in Gdt1p/TMEM165 deficient cells result from a defect in Golgi manganese homeostasis. Hum. Mol. Genet. 2016, 25, 1489–1500. [Google Scholar] [CrossRef] [Green Version]
- Dulary, E.; Potelle, S.; Legrand, D.; Foulquier, F. TMEM165 deficiencies in Congenital Disorders of Glycosylation type II (CDG-II): Clues and evidences for roles of the protein in Golgi functions and ion homeostasis. Tissue Cell 2017, 49, 150–156. [Google Scholar] [CrossRef]
- Clark, C.R.; Maile, M.; Blaney, P.; Hellweg, S.R.; Strauss, A.; Durose, W.; Priya, S.; Habicht, J.; Burns, M.B.; Blekhman, R.; et al. Transposon mutagenesis screen in mice identifies TM9SF2 as a novel colorectal cancer oncogene. Sci. Rep. 2018, 8, 15327. [Google Scholar] [CrossRef] [PubMed]
- Schimmoller, F.; Diaz, E.; Muhlbauer, B.; Pfeffer, S.R. Characterization of a 76 kDa endosomal, multispanning membrane protein that is highly conserved throughout evolution. Gene 1998, 216, 311–318. [Google Scholar] [CrossRef]
- Bergeret, E.; Perrin, J.; Williams, M.; Grunwald, D.; Engel, E.; Thevenon, D.; Taillebourg, E.; Bruckert, F.; Cosson, P.; Fauvarque, M.O. TM9SF4 is required for Drosophila cellular immunity via cell adhesion and phagocytosis. J. Cell Sci. 2008, 121, 3325–3334. [Google Scholar] [CrossRef] [Green Version]
- Cornillon, S.; Pech, E.; Benghezal, M.; Ravanel, K.; Gaynor, E.; Letourneur, F.; Bruckert, F.; Cosson, P. Phg1p is a nine-transmembrane protein superfamily member involved in dictyostelium adhesion and phagocytosis. J. Biol. Chem. 2000, 275, 34287–34292. [Google Scholar] [CrossRef] [Green Version]
- Pastan, I.; Hassan, R.; Fitzgerald, D.J.; Kreitman, R.J. Immunotoxin therapy of cancer. Nat. Rev. Cancer 2006, 6, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Antignani, A.; Fitzgerald, D. Immunotoxins: The role of the toxin. Toxins 2013, 5, 1486–1502. [Google Scholar] [CrossRef] [Green Version]
- Oldham, R.A.A.; Faber, M.L.; Keppel, T.R.; Buchberger, A.R.; Waas, M.; Hari, P.; Gundry, R.L.; Medin, J.A. Discovery and validation of surface N-glycoproteins in MM cell lines and patient samples uncovers immunotherapy targets. J. Immunother. Cancer 2020, 8. [Google Scholar] [CrossRef]
- Fanale, M.A.; Hamlin, P.A.; Park, S.I.; Persky, D.O.; Higgins, J.P.; Burnett, C.; Dabovic, K.; Poma, E.; Sarapa, N.; Younes, A. Safety and efficacy of anti-CD20 immunotoxin MT-3724 in relapsed/refractory (R/R) B-cell non-Hodgkin lymphoma (NHL) in a phase I study. J. Clin. Oncol. 2018, 36, 7580. [Google Scholar] [CrossRef]
- Higgins, J.P.; Iberg, A.; Howard, C.; Willert, E. Abstract 2060: Combination of CD20 targeted engineered toxin body, MT-3724, with chemotherapy or IMiDs for the treatment of non Hodgkin’s lymphoma. Cancer Res. 2019, 79, 2060. [Google Scholar] [CrossRef]
- Persky, D.O.; Musteata, V.; Zodelava, M.; Perekhrestenko, T.; Diaz, A.E.; Guthrie, T.H., Jr.; Strack, T.; Burnett, C.; Wilson, S.; Baetz, T. A Phase 2 Study of MT-3724 to Evaluate Safety, Pharmacodynamics and Efficacy of MT-3724 for the Treatment of Patients with Relapsed or Refractory Diffuse Large B-Cell Lymphoma. Blood 2019, 134, 5324. [Google Scholar] [CrossRef]
- Erin, K.; Willert, G.L.R.; Jack, P.; Higgins, J.L.; Lee, J.; Syed, S.; Zhang, Y.; Tavares, D.; Lublinsky, A.; Chattopadhyay, N.; et al. TAK-169, an exceptionally potent CD38 targeted engineered toxin body, as a novel direct cell kill approach for the treatment of multiple myeloma [abstract]. In Proceedings of the American Association for Cancer Research Annual Meeting 2019, Atlanta, GA, USA, 29 March–3 April 2019; AACR: Philadelphia, PA, USA, 2019; Volume 79. [Google Scholar]
- Kumar, S.K.; Cornell, R.F.; Landgren, O.; Ailawadhi, S.; Higgins, J.P.; Willert, E.K.; Waltzman, R.; Lin, J.; Zhang, Y.; Lublinsky, A.R.; et al. A Phase 1 First-in-Human Study of the Anti-CD38 Dimeric Fusion Protein TAK-169 for the Treatment of Patients (pts) with Relapsed or Refractory Multiple Myeloma (RRMM) Who Are Proteasome Inhibitor (PI)- and Immunomodulatory Drug (IMiD)-Refractory, Including Pts Relapsed/Refractory (R/R) or Naïve to Daratumumab (dara). Blood 2019, 134, 1867. [Google Scholar] [CrossRef]
- Higgins, J.P.; Sarkar, A.; Williams, E.T.; Iberg, A.; Waltzman, R.; Willert, E.K. Abstract P1-18-35: MT-5111, a novel HER2 targeting engineered toxin body, under clinical development to overcome mechanisms of resistance to existing HER2 targeted therapies. Cancer Res. 2020, 80, P1-18-35–P11-18-35. [Google Scholar] [CrossRef]
- Waltzman, R.J.; Sarkar, A.; Williams, E.T.; Iberg, A.T.; Higgins, J.T.; Willert, E.K. MT-5111: A novel HER2 targeting engineered toxin body in clinical development. J. Clin. Oncol. 2020, 38, 433. [Google Scholar] [CrossRef]
- Waltzman, R.J.; Brieschke, B.; LeMar, S.; Dekker, J.; Cornelison, G.; Robinson, G.L.; Sarkar, A.; Zhao, J.; Iberg, A.T.; Ramos, H.J.; et al. PD-L1 targeted engineered toxin body provides direct cytotoxicity and T-cell mediated tumor targeting. J. Clin. Oncol. 2020, 38, 12. [Google Scholar] [CrossRef]
- Ramos, H.J.; Brieschke, B.; LeMar, S.; Dekker, J.D.; Iberg, A.; Robinson, G.L.; Sarkar, A.; Anand, B.; Singh, M.M.; Zhao, J.; et al. Abstract 3366: In vivo efficacy of a PD-L1 targeted, antigen seeding engineered toxin body. Cancer Res. 2020, 80, 3366. [Google Scholar] [CrossRef]
- Lee, R.S.; Tartour, E.; van der Bruggen, P.; Vantomme, V.; Joyeux, I.; Goud, B.; Fridman, W.H.; Johannes, L. Major histocompatibility complex class I presentation of exogenous soluble tumor antigen fused to the B-fragment of Shiga toxin. Eur. J. Immunol. 1998, 28, 2726–2737. [Google Scholar] [CrossRef]
- Haicheur, N.; Bismuth, E.; Bosset, S.; Adotevi, O.; Warnier, G.; Lacabanne, V.; Regnault, A.; Desaymard, C.; Amigorena, S.; Ricciardi-Castagnoli, P.; et al. The B subunit of Shiga toxin fused to a tumor antigen elicits CTL and targets dendritic cells to allow MHC class I-restricted presentation of peptides derived from exogenous antigens. J. Immunol. 2000, 165, 3301–3308. [Google Scholar] [CrossRef] [Green Version]
- Haicheur, N.; Benchetrit, F.; Amessou, M.; Leclerc, C.; Falguieres, T.; Fayolle, C.; Bismuth, E.; Fridman, W.H.; Johannes, L.; Tartour, E. The B subunit of Shiga toxin coupled to full-size antigenic protein elicits humoral and cell-mediated immune responses associated with a Th1-dominant polarization. Int. Immunol. 2003, 15, 1161–1171. [Google Scholar] [CrossRef] [Green Version]
- Choi, N.W.; Estes, M.K.; Langridge, W.H. Oral immunization with a shiga toxin B subunit: Rotavirus NSP4(90) fusion protein protects mice against gastroenteritis. Vaccine 2005, 23, 5168–5176. [Google Scholar] [CrossRef]
- Vingert, B.; Adotevi, O.; Patin, D.; Jung, S.; Shrikant, P.; Freyburger, L.; Eppolito, C.; Sapoznikov, A.; Amessou, M.; Quintin-Colonna, F.; et al. The Shiga toxin B-subunit targets antigen in vivo to dendritic cells and elicits anti-tumor immunity. Eur. J. Immunol. 2006, 36, 1124–1135. [Google Scholar] [CrossRef]
- Kordbacheh, E.; Nazarian, S.; Hajizadeh, A.; Fasihi-Ramandi, M.; Fathi, J. Recombinant HcpA-EspA-Tir-Stx2B chimeric protein induces immunity against attachment and toxicity of Escherichia coli O157:H7. Microb. Pathog. 2019, 129, 176–182. [Google Scholar] [CrossRef]
- Yanina, H.; Romina, P.; Lucas, B.; Constanza, L.; Luciana, M.; Berengeno Andrea, L.; Mariana, C.; Ortega, H.; Goldbaum Fernando, A.; Santiago, S.; et al. Preclinical Studies of NEAST (Neutralizing Equine Anti-Shiga To xin): A Potential Treatment for Prevention of Stec-Hus. Int. J. Drug Dev. Res. 2019, 11. [Google Scholar] [CrossRef]
- LaCasse, E.C.; Bray, M.R.; Patterson, B.; Lim, W.M.; Perampalam, S.; Radvanyi, L.G.; Keating, A.; Stewart, A.K.; Buckstein, R.; Sandhu, J.S.; et al. Shiga-like toxin-1 receptor on human breast cancer, lymphoma, and myeloma and absence from CD34(+) hematopoietic stem cells: Implications for ex vivo tumor purging and autologous stem cell transplantation. Blood 1999, 94, 2901–2910. [Google Scholar]
- Johansson, D.; Kosovac, E.; Moharer, J.; Ljuslinder, I.; Brannstrom, T.; Johansson, A.; Behnam-Motlagh, P. Expression of verotoxin-1 receptor Gb3 in breast cancer tissue and verotoxin-1 signal transduction to apoptosis. BMC Cancer 2009, 9, 67. [Google Scholar] [CrossRef] [Green Version]
- Stimmer, L.; Dehay, S.; Nemati, F.; Massonnet, G.; Richon, S.; Decaudin, D.; Klijanienko, J.; Johannes, L. Human breast cancer and lymph node metastases express Gb3 and can be targeted by STxB-vectorized chemotherapeutic compounds. BMC Cancer 2014, 14, 916. [Google Scholar] [CrossRef] [PubMed]
- Arab, S.; Russel, E.; Chapman, W.B.; Rosen, B.; Lingwood, C.A. Expression of the verotoxin receptor glycolipid, globotriaosylceramide, in ovarian hyperplasias. Oncol. Res. 1997, 9, 553–563. [Google Scholar] [PubMed]
- Heath-Engel, H.M.; Lingwood, C.A. Verotoxin sensitivity of ECV304 cells in vitro and in vivo in a xenograft tumour model: VT1 as a tumour neovascular marker. Angiogenesis 2003, 6, 129–141. [Google Scholar] [CrossRef]
- Maak, M.; Nitsche, U.; Keller, L.; Wolf, P.; Sarr, M.; Thiebaud, M.; Rosenberg, R.; Langer, R.; Kleeff, J.; Friess, H.; et al. Tumor-specific targeting of pancreatic cancer with Shiga toxin B-subunit. Mol. Cancer Ther. 2011, 10, 1918–1928. [Google Scholar] [CrossRef] [Green Version]
- Kovbasnjuk, O.; Mourtazina, R.; Baibakov, B.; Wang, T.; Elowsky, C.; Choti, M.A.; Kane, A.; Donowitz, M. The glycosphingolipid globotriaosylceramide in the metastatic transformation of colon cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 19087–19092. [Google Scholar] [CrossRef] [Green Version]
- Falguieres, T.; Maak, M.; von Weyhern, C.; Sarr, M.; Sastre, X.; Poupon, M.F.; Robine, S.; Johannes, L.; Janssen, K.P. Human colorectal tumors and metastases express Gb3 and can be targeted by an intestinal pathogen-based delivery tool. Mol. Cancer Ther. 2008, 7, 2498–2508. [Google Scholar] [CrossRef] [Green Version]
- Farkas-Himsley, H.; Hill, R.; Rosen, B.; Arab, S.; Lingwood, C.A. The bacterial colicin active against tumor cells in vitro and in vivo is verotoxin 1. Proc. Natl. Acad. Sci. USA 1995, 92, 6996–7000. [Google Scholar] [CrossRef] [Green Version]
- Geyer, P.E.; Maak, M.; Nitsche, U.; Perl, M.; Novotny, A.; Slotta-Huspenina, J.; Dransart, E.; Holtorf, A.; Johannes, L.; Janssen, K.P. Gastric Adenocarcinomas Express the Glycosphingolipid Gb3/CD77: Targeting of Gastric Cancer Cells with Shiga Toxin B-Subunit. Mol. Cancer Ther. 2016, 15, 1008–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salhia, B.; Rutka, J.T.; Lingwood, C.; Nutikka, A.; Van Furth, W.R. The treatment of malignant meningioma with verotoxin. Neoplasia 2002, 4, 304–311. [Google Scholar] [CrossRef] [Green Version]
- Couture, O.; Dransart, E.; Dehay, S.; Nemati, F.; Decaudin, D.; Johannes, L.; Tanter, M. Tumor delivery of ultrasound contrast agents using Shiga toxin B subunit. Mol. Imaging 2011, 10, 135–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.Y.; Tang, J.; He, E.H.; Li, Y.M.; Zhou, Y.; Zhang, X.; Chen, G. Clinical utility of three-dimensional contrast-enhanced ultrasound in the differentiation between noninvasive and invasive neoplasms of urinary bladder. Eur. J. Radiol. 2012, 81, 2936–2942. [Google Scholar] [CrossRef]
- Nicolau, C.; Bunesch, L.; Peri, L.; Salvador, R.; Corral, J.M.; Mallofre, C.; Sebastia, C. Accuracy of contrast-enhanced ultrasound in the detection of bladder cancer. Br. J. Radiol. 2011, 84, 1091–1099. [Google Scholar] [CrossRef] [Green Version]
- Tarrago-Trani, M.T.; Jiang, S.; Harich, K.C.; Storrie, B. Shiga-like toxin subunit B (SLTB)-enhanced delivery of chlorin e6 (Ce6) improves cell killing. Photochem. Photobiol. 2006, 82, 527–537. [Google Scholar] [CrossRef]
- El Alaoui, A.; Schmidt, F.; Amessou, M.; Sarr, M.; Decaudin, D.; Florent, J.C.; Johannes, L. Shiga toxin-mediated retrograde delivery of a topoisomerase I inhibitor prodrug. Angew. Chem. Int. Ed. Engl. 2007, 46, 6469–6472. [Google Scholar] [CrossRef]
- El Alaoui, A.; Schmidt, F.; Sarr, M.; Decaudin, D.; Florent, J.C.; Johannes, L. Synthesis and properties of a mitochondrial peripheral benzodiazepine receptor conjugate. ChemMedChem 2008, 3, 1687–1695. [Google Scholar] [CrossRef]
- Batisse, C.; Dransart, E.; Ait Sarkouh, R.; Brulle, L.; Bai, S.K.; Godefroy, S.; Johannes, L.; Schmidt, F. A new delivery system for auristatin in STxB-drug conjugate therapy. Eur. J. Med. Chem. 2015, 95, 483–491. [Google Scholar] [CrossRef]
- Kostova, V.; Dransart, E.; Azoulay, M.; Brulle, L.; Bai, S.K.; Florent, J.C.; Johannes, L.; Schmidt, F. Targeted Shiga toxin-drug conjugates prepared via Cu-free click chemistry. Bioorg. Med. Chem. 2015, 23, 7150–7157. [Google Scholar] [CrossRef]
- Ryou, J.H.; Sohn, Y.K.; Hwang, D.E.; Kim, H.S. Shiga-like toxin-based high-efficiency and receptor-specific intracellular delivery system for a protein. Biochem. Biophys. Res. Commun. 2015, 464, 1282–1289. [Google Scholar] [CrossRef]
- Mohammed, A.F.; Abdul-Wahid, A.; Huang, E.H.; Bolewska-Pedyczak, E.; Cydzik, M.; Broad, A.E.; Gariepy, J. The Pseudomonas aeruginosa exotoxin A translocation domain facilitates the routing of CPP-protein cargos to the cytosol of eukaryotic cells. J. Control. Release 2012, 164, 58–64. [Google Scholar] [CrossRef]
- Ryou, J.H.; Sohn, Y.K.; Hwang, D.E.; Park, W.Y.; Kim, N.; Heo, W.D.; Kim, M.Y.; Kim, H.S. Engineering of bacterial exotoxins for highly efficient and receptor-specific intracellular delivery of diverse cargos. Biotechnol. Bioeng. 2016, 113, 1639–1646. [Google Scholar] [CrossRef] [PubMed]
- Ryou, J.H.; Sohn, Y.K.; Kim, D.G.; Kyeong, H.H.; Kim, H.S. Engineering and cytosolic delivery of a native regulatory protein and its variants for modulation of ERK2 signaling pathway. Biotechnol. Bioeng. 2018, 115, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Schmit, N.E.; Neopane, K.; Hantschel, O. Targeted Protein Degradation through Cytosolic Delivery of Monobody Binders Using Bacterial Toxins. ACS Chem. Biol. 2019, 14, 916–924. [Google Scholar] [CrossRef]
- Du, J.; Jin, J.; Yan, M.; Lu, Y. Synthetic nanocarriers for intracellular protein delivery. Curr. Drug Metab. 2012, 13, 82–92. [Google Scholar] [CrossRef]
- Frank, C.; Werber, D.; Cramer, J.P.; Askar, M.; Faber, M.; an der Heiden, M.; Bernard, H.; Fruth, A.; Prager, R.; Spode, A.; et al. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N. Engl. J. Med. 2011, 365, 1771–1780. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Tian, S.; Thaker, H.; Dong, M. Shiga Toxins: An Update on Host Factors and Biomedical Applications. Toxins 2021, 13, 222. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13030222
Liu Y, Tian S, Thaker H, Dong M. Shiga Toxins: An Update on Host Factors and Biomedical Applications. Toxins. 2021; 13(3):222. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13030222
Chicago/Turabian StyleLiu, Yang, Songhai Tian, Hatim Thaker, and Min Dong. 2021. "Shiga Toxins: An Update on Host Factors and Biomedical Applications" Toxins 13, no. 3: 222. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13030222




