Short Linear Motifs Characterizing Snake Venom and Mammalian Phospholipases A2
Abstract
:1. Introduction
2. Results
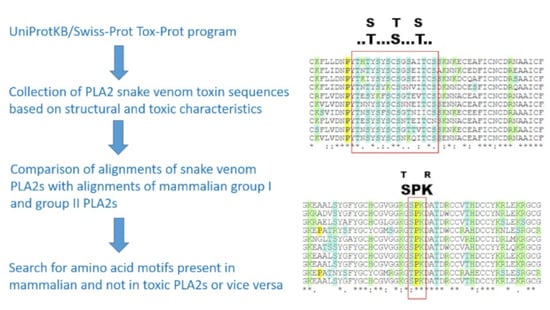
2.1. Sequence Alignment Comparison
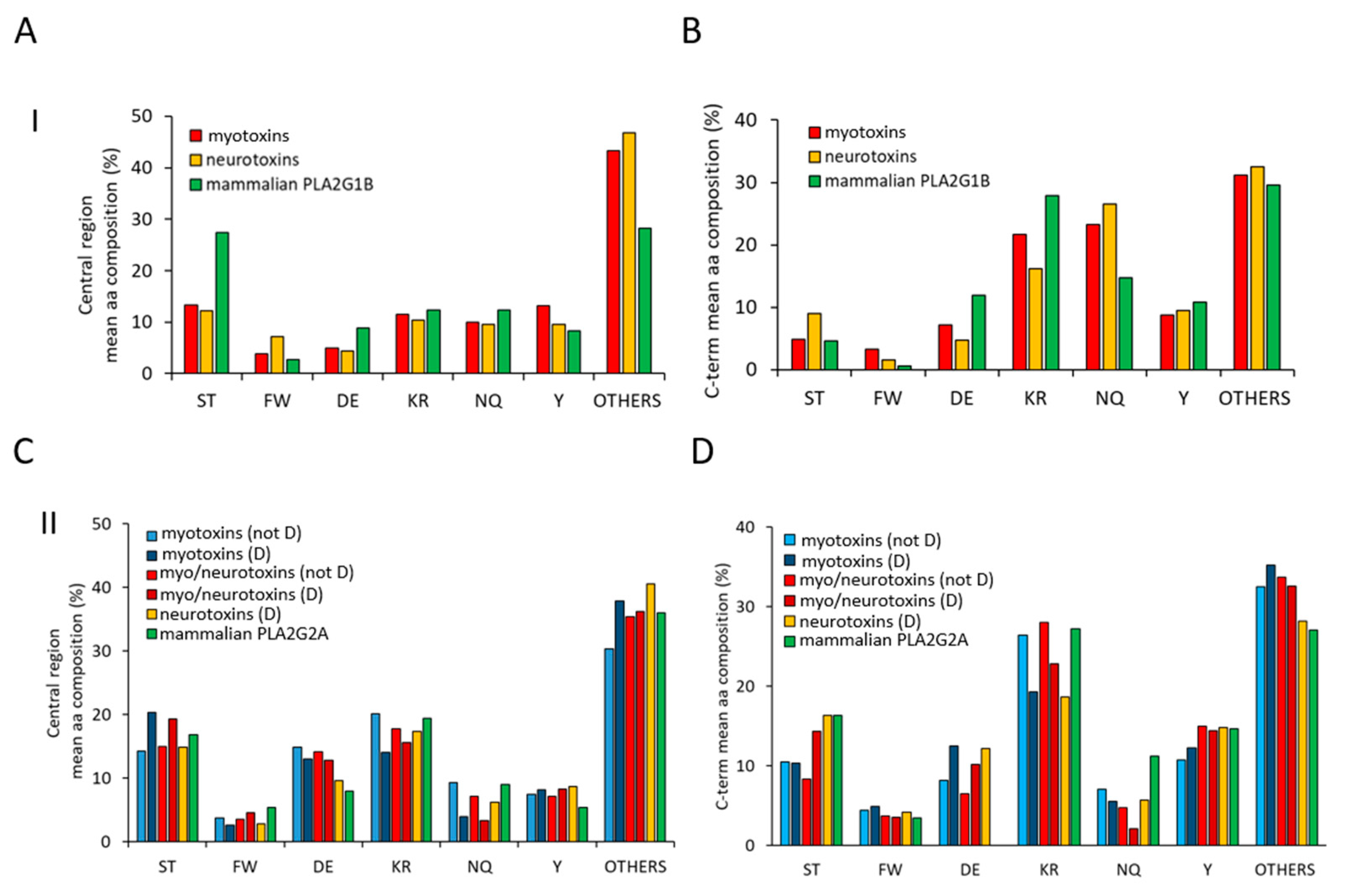
2.2. Amino Acid Composition Analysis of the Central and C-Terminal Regions
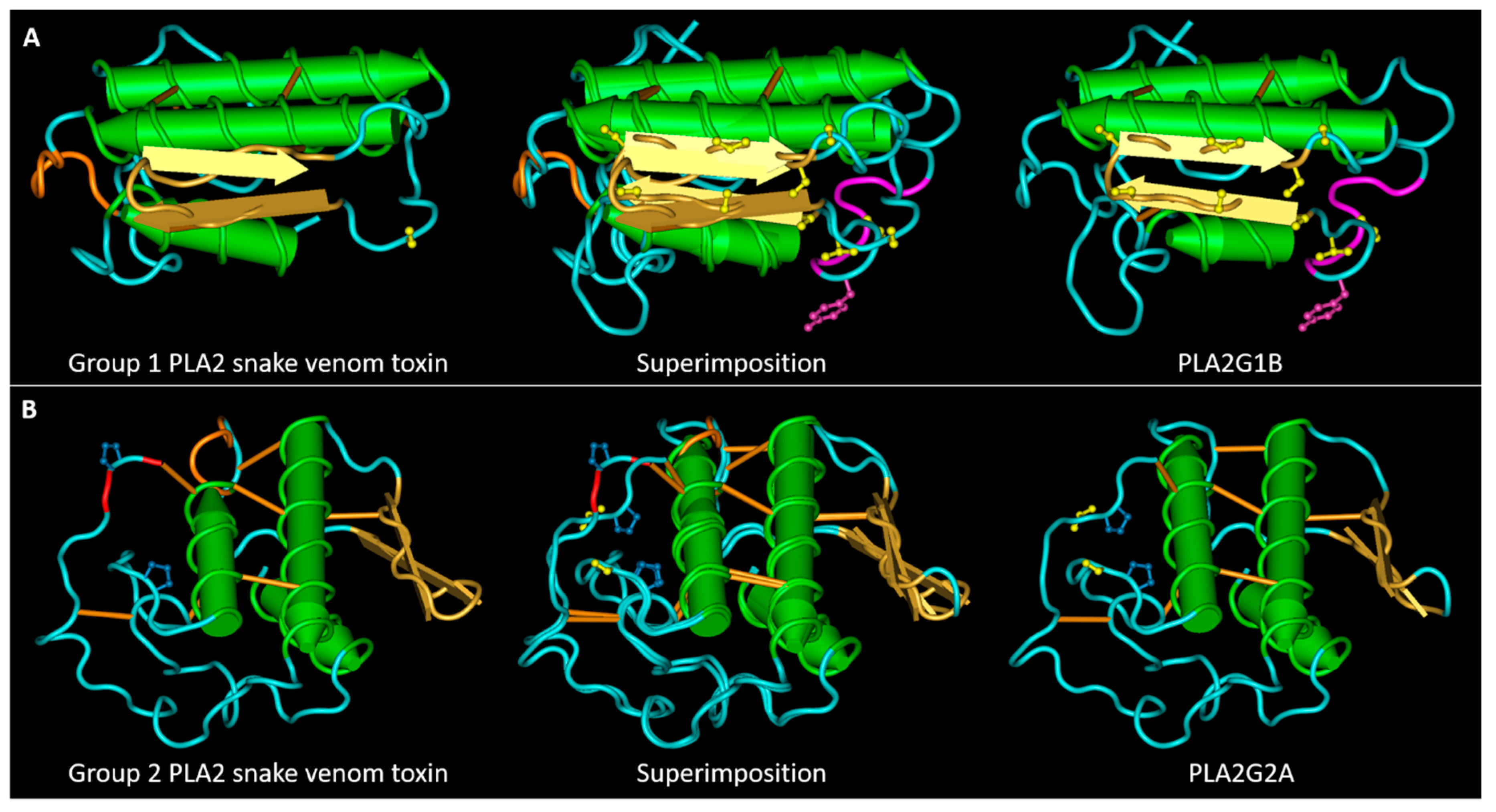
2.3. SLiMs Conserved in Toxins and Not in Mammalian PLA2s or Vice Versa
| Motif Description | ELM Class | Sequence Section Start–End | Fraction of Proteins Containing the Motif in the Specified Section | ||
|---|---|---|---|---|---|
| Neuro-Myotoxins | Neurotoxins | PLA2G1B | |||
| N-terminal | DEG_Nend_UBRbox_3 | 1 | 7/7 | 14/14 | 0/10 |
| NEK2 phosph. site | MOD_NEK2_1 | 31–36 | 0/7 | 0/14 | 9/10 |
| I-BAR binding site | LIG_IBAR_NPY_1 | 67–69 | 0/7 | 0/14 | 10/10 |
| SH2 binding site | LIG_SH2_STAP1 | 69–73 | 0/7 | 0/14 | 10/10 |
| GSK3 phosph. sites * | MOD_GSK3_1 | 67–80 | 0/7 | 1/14 | 10/10 |
| SH2 binding site | LIG_SH2_NCK_1 | 105–109 | 3/7 | 6/14 | 0/10 |
| PTB binding sites | LIG_PTB_Apo_2LIG_PTB_Phospho_1 | 104–111 | 1/7 | 9/14 | 0/10 |
| Motif Description | ELM Class | Sequence Section Start–End | Fraction of Proteins Containing the Motif in the Specified Section | |||||
|---|---|---|---|---|---|---|---|---|
| Myotoxins | Neuro-Myotoxins | Neurotoxins | PLA2G2A | |||||
| NOT D49 | D49 | NOT D49 | D49 | D49 | ||||
| Pin1 site, and PDK and CDK phosph. sites | DOC_WW_Pin1_4 MOD_ProDKin_1 MOD_CDK_SPK_2 | 35–37 | 0/25 | 0/14 | 1/5 | 1/9 | 4/12 | 10/10 |
| SH2 binding site | LIG_SH2_CRK | 51–55 | 23/25 | 9/14 | 3/5 | 6/9 | 9/12 | 0/10 |
| PKA phosph. site | MOD_PKA_1 | 52–56 | 23/25 | 3/14 | 3/5 | 1/9 | 0/12 | 0/10 |
| FHA binding site | LIG_FHA_1 | 59–65 | 0/25 | 0/14 | 0/5 | 2/9 | 4/12 | 10/10 |
| Pin1 site and CDK phosph. site | DOC_WW_Pin1_4 MOD_CDK_SPK_2 | 119–121 | 0/25 | 0/14 | 0/5 | 0/9 | 0/12 | 7/10 |
| PDZ binding site | LIG_PDZ_Class_3 | 119–121 | 23/25 | 9/14 | 5/5 | 5/9 | 10/12 | 0/10 |
| ELM Class Identifier | Regular Expression | Interaction Partner(s) | Examples of Proteins * Containing the SLiM |
|---|---|---|---|
| DEG_Nend_ UBRbox_3 | ^M{0,1}([NQ]) | Aminohydrolases for deamidation | - |
| DOC_WW_Pin1_4 | ...([ST])P. | Peptidyl prolyl isomerase 1 | Many |
| LIG_FHA_1 | ..(T)..[ILV]. | Proteins containing the forkhead-associated domain, e.g., TIFA, TIFAB, AGGF1 | Kinesin interactors, TIFA |
| LIG_IBAR_NPY_1 | NPY | I-BAR domain-containing proteins, involved in membrane dynamic | The bacterial protein Tir, SHANK2 |
| LIG_PDZ_Class_3 | ...[DE].[ACVILF]$ | PDZ containing proteins, PDZ domains also bind to phospholipid headgroups | Many PDZ ligands are membrane proteins |
| LIG_PTB_Apo_2 LIG_PTB_Phospho_1 | (.[^P].NP.[FY].)|(.[ILVMFY].N..[FY].) (.[^P].NP.(Y))|(.[ILVMFY].N..(Y)) | Proteins containing phosphotyrosine binding (PTB) domains, e.g., insulin receptor substrate 1 (IRS-1) | Integrins, LRP1 |
| LIG_SH2_CRK | (Y)[^EPILVFYW][^HDEW][PLIV][^DEW] | Proteins containing SH2 domain of the CRK family | Transmembrane receptors, Phospholipase C-gamma-1 |
| LIG_SH2_NCK_1 | (Y)[DESTNA][^GWFY][VPAI][DENQSTAGYFP] | Proteins containing SH2 domain of the NCK family | Transmembrane proteins, adapter protein docking 1 |
| LIG_SH2_STAP1 | (Y)[DESTA][^GP][^GP][ILVFMWYA] | Proteins containing SH2 domain of the STAP1 family | Transmembrane proteins, lipid phosphatase |
| MOD_CDK_SPK_2 | ...([ST])P[RK] | Cyclin-dependent kinases, proline directed kinase | Proteins involved in different biochemical pathways |
| MOD_GSK3_1 | ...([ST])...[ST] | Glycogen synthase kinase 3, needs priming, inhibitory | |
| MOD_NEK2_1 | [FLM][^P][^P]([ST])[^DEP][^DE] | Never in mitosis A (NimA)-related kinases | |
| MOD_PKA_1 | [RK][RK].([ST])[^P].. | cAMP-dependent protein kinase A, basophilic kinase | |
| MOD_ProDKin_1 | ...([ST])P.. | MAP Kinase, proline directed kinase |
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Sequence Collection and Alignment
5.2. Amino-Acidic Composition Analysis of the β-Sheet Containing Region and C-Terminal Stretch
5.3. Short Linear Motifs Identification
5.4. Tri-Dimensional Structure Representations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dennis, E. Diversity of group types, regulation, and function of phospholipase A2. J. Biol. Chem. 1994, 269, 13057–13060. [Google Scholar] [CrossRef]
- Filkin, S.Y.; Lipkin, A.V.; Fedorov, A.N. Phospholipase Superfamily: Structure, Functions, and Biotechnological Applications. Biochemistry 2020, 85, 177–195. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.E.; Dennis, E.A. Phospholipase A2 biochemistry. Cardiovasc. Drugs Ther. 2008, 23, 49–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, M.; Taketomi, Y.; Sato, H.; Yamamoto, K. Secreted phospholipase A2 revisited. J. Biochem. 2011, 150, 233–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaloske, R.H.; Dennis, E.A. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta 2006, 1761, 1246–1259. [Google Scholar] [CrossRef]
- Dennis, E.A.; Cao, J.; Hsu, Y.-H.; Magrioti, V.; Kokotos, G. Phospholipase A2Enzymes: Physical Structure, Biological Function, Disease Implication, Chemical Inhibition, and Therapeutic Intervention. Chem. Rev. 2011, 111, 6130–6185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, M.; Sato, H.; Miki, Y.; Yamamoto, K.; Taketomi, Y. A new era of secreted phospholipase A₂. J. Lipid Res. 2015, 56, 1248–1261. [Google Scholar] [CrossRef] [Green Version]
- Singer, A.G.; Ghomashchi, F.; Le Calvez, C.; Bollinger, J.; Bezzine, S.; Rouault, M.; Sadilek, M.; Nguyen, E.; Lazdunski, M.; Lambeau, G.; et al. Interfacial Kinetic and Binding Properties of the Complete Set of Human and Mouse Groups I, II, V, X, and XII Secreted Phospholipases A2. J. Biol. Chem. 2002, 277, 48535–48549. [Google Scholar] [CrossRef] [Green Version]
- Murakami, M.; Sato, H.; Taketomi, Y. Updating Phospholipase A(2) Biology. Biomolecules 2020, 10, 1457. [Google Scholar] [CrossRef]
- El-Aziz, T.M.A.; Soares, A.G.; Stockand, J.D. Snake Venoms in Drug Discovery: Valuable Therapeutic Tools for Life Saving. Toxins 2019, 11, 564. [Google Scholar] [CrossRef] [Green Version]
- Tasoulis, T.; Isbister, G.K. A Review and Database of Snake Venom Proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef] [Green Version]
- Tonello, F.; Rigoni, M. Cellular Mechanisms of Action of Snake Phospholipase A2 Toxins. In Snake Venoms; Inagaki, H., Vogel, C.-W., Mukherjee, A.K., Rahmy, T.R., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 49–65. ISBN 978-94-007-6410-1. [Google Scholar]
- Ferraz, C.R.; Arrahman, A.; Xie, C.; Casewell, N.R.; Lewis, R.J.; Kool, J.; Cardoso, F.C. Multifunctional Toxins in Snake Venoms and Therapeutic Implications: From Pain to Hemorrhage and Necrosis. Front. Ecol. Evol. 2019, 7, 218. [Google Scholar] [CrossRef] [Green Version]
- Arni, R.K.; Ward, R.J. Phospholipase A2—A structural review. Toxicon 1996, 34, 827–841. [Google Scholar] [CrossRef]
- Nevalainen, T.J.; Cardoso, J.C.R.; Riikonen, P.T. Conserved domains and evolution of secreted phospholipases A2. FEBS J. 2011, 279, 636–649. [Google Scholar] [CrossRef] [Green Version]
- Lomonte, B.; Gutiérrez, J.M. Phospholipases A2 from viperidae snake venoms: How do they induce skeletal muscle damage? Acta Chim. Slov. 2011, 58, 647–659. [Google Scholar]
- Lomonte, B.; Rangel, J. Snake venom Lys49 myotoxins: From phospholipases A2 to non-enzymatic membrane disruptors. Toxicon 2012, 60, 520–530. [Google Scholar] [CrossRef]
- Murakami, M.; Taketomi, Y.; Miki, Y.; Sato, H.; Yamamoto, K.; Lambeau, G. Emerging roles of secreted phospholipase A2 enzymes: The 3rd edition. Biochimie 2014, 107, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Yarla, N.S.; Azad, R.; Basha, M.; Rajack, A.; Kaladhar, D.S.V.G.K.; Allam, B.K.; Pragada, R.R.; Singh, K.N.; Kumari, K.S.; Pallu, R.; et al. 5-Lipoxygenase and cyclooxygenase inhibitory dammarane triterpenoid 1 from Borassus flabellifer seed coat inhibits tumor necrosis factor-α secretion in LPSInduced THP-1 human monocytes and induces apoptosis in MIA PaCa-2 pancreatic cancer cells. Anticancer. Agents Med. Chem. 2015, 15, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Lambeau, G.; Gelb, M.H. Biochemistry and Physiology of Mammalian Secreted Phospholipases A2. Annu. Rev. Biochem. 2008, 77, 495–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brglez, V.; Lambeau, G.; Petan, T. Secreted phospholipases A2 in cancer: Diverse mechanisms of action. Biochimie 2014, 107, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Jungo, F.; Bougueleret, L.; Xenarios, I.; Poux, S. The UniProtKB/Swiss-Prot Tox-Prot program: A central hub of integrated venom protein data. Toxicon 2012, 60, 551–557. [Google Scholar] [CrossRef] [Green Version]
- Gouw, M.; Sámano-Sánchez, H.; Van Roey, K.; Diella, F.; Gibson, T.J.; Dinkel, H. Exploring Short Linear Motifs Using the ELM Database and Tools. Curr. Protoc. Bioinform. 2017, 58, 8.22.1–8.22.35. [Google Scholar] [CrossRef]
- Kumar, M.; Gouw, M.; Michael, S.; Sámano-Sánchez, H.; Pancsa, R.; Glavina, J.; Diakogianni, A.; Valverde, J.A.; Bukirova, D.; Čalyševa, J.; et al. ELM—the eukaryotic linear motif resource in 2020. Nucleic Acids Res. 2019, 48, D296–D306. [Google Scholar] [CrossRef] [Green Version]
- Thalmann, R.; Ignatova, E.; Kachar, B.; Ornitz, D.M.; Thalmann, I. Development and maintenance of otoconia: Biochemical considerations. Ann. N. Y. Acad. Sci. 2001, 942, 162–178. [Google Scholar] [CrossRef]
- Massimino, M.L.; Simonato, M.; Spolaore, B.; Franchin, C.; Arrigoni, G.; Marin, O.; Monturiol-Gross, L.; Fernández, J.; Lomonte, B.; Tonello, F. Cell surface nucleolin interacts with and internalizes Bothrops asper Lys49 phospholipase A2 and mediates its toxic activity. Sci. Rep. 2018, 8, 10619. [Google Scholar] [CrossRef] [Green Version]
- Spolaore, B.; Fernández, J.; Lomonte, B.; Massimino, M.L.; Tonello, F. Enzymatic labelling of snake venom phospholipase A2 toxins. Toxicon 2019, 170, 99–107. [Google Scholar] [CrossRef]
- Šribar, J.; Kovačič, L.; Oberčkal, J.; Ivanušec, A.; Petan, T.; Fox, J.W.; Križaj, I. The neurotoxic secreted phospholipase A. Sci. Rep. 2019, 9, 283. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Kambe, T.; Shimbara, S.; Kudo, I. Functional Coupling Between Various Phospholipase A2s and Cyclooxygenases in Immediate and Delayed Prostanoid Biosynthetic Pathways. J. Biol. Chem. 1999, 274, 3103–3115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigoni, M.; Paoli, M.; Milanesi, E.; Caccin, P.; Rasola, A.; Bernardi, P.; Montecucco, C. Snake Phospholipase A2 Neurotoxins Enter Neurons, Bind Specifically to Mitochondria, and Open Their Transition Pores. J. Biol. Chem. 2008, 283, 34013–34020. [Google Scholar] [CrossRef] [Green Version]
- Tasaki, T.; Sriram, S.M.; Park, K.S.; Kwon, Y.T. The N-End Rule Pathway. Annu. Rev. Biochem. 2012, 81, 261–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, A.P.D.P.D.; Issayama, L.K.K.; Pavan, I.C.B.B.; Silva, F.R.R.; Melo-Hanchuk, T.D.D.; Simabuco, F.M.M.; Kobarg, J. Checking NEKs: Overcoming a Bottleneck in Human Diseases. Molecules 2020, 25, 1778. [Google Scholar] [CrossRef]
- Sámano-Sánchez, H.; Gibson, T.J. Mimicry of Short Linear Motifs by Bacterial Pathogens: A Drugging Opportunity. Trends Biochem. Sci. 2020, 45, 526–544. [Google Scholar] [CrossRef]
- Masuhara, M.; Nagao, K.; Nishikawa, M.; Sasaki, M.; Yoshimura, A.; Osawa, M. Molecular Cloning of Murine STAP-1, the Stem-Cell-Specific Adaptor Protein Containing PH and SH2 Domains. Biochem. Biophys. Res. Commun. 2000, 268, 697–703. [Google Scholar] [CrossRef]
- Cesaro, L.; Pinna, L.A.; Salvi, M. A Comparative Analysis and Review of lysyl Residues Affected by Posttranslational Modifications. Curr. Genom. 2015, 16, 128–138. [Google Scholar] [CrossRef] [Green Version]
- Hoffmeister, L.; Diekmann, M.; Brand, K.; Huber, R. GSK3: A Kinase Balancing Promotion and Resolution of Inflammation. Cells 2020, 9, 820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaqaman, K.; Ditlev, J.A. Biomolecular condensates in membrane receptor signaling. Curr. Opin. Cell Biol. 2021, 69, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Case, L.B.; Ditlev, J.A.; Rosen, M.K. Regulation of Transmembrane Signaling by Phase Separation. Annu. Rev. Biophys. 2019, 48, 465–494. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, Y.; Matsunaga, Y.; Yamamotoya, T.; Ueda, K.; Inoue, Y.; Mori, K.; Sakoda, H.; Fujishiro, M.; Ono, H.; Kushiyama, A.; et al. Physiological and Pathogenic Roles of Prolyl Isomerase Pin1 in Metabolic Regulations via Multiple Signal Transduction Pathway Modulations. Int. J. Mol. Sci. 2016, 17, 1495. [Google Scholar] [CrossRef]
- Chioato, L.; Aragão, E.A.; Ferreira, T.L.; De Medeiros, A.I.; Faccioli, L.H.; Ward, R.J. Mapping of the structural determinants of artificial and biological membrane damaging activities of a Lys49 phospholipase A2 by scanning alanine mutagenesis. Biochim. Biophys. Acta 2007, 1768, 1247–1257. [Google Scholar] [CrossRef] [Green Version]
- Rogne, M.; Taskén, K. Compartmentalization of cAMP Signaling in Adipogenesis, Lipogenesis, and Lipolysis. Horm. Metab. Res. 2014, 46, 833–840. [Google Scholar] [CrossRef]
- Niederkorn, M.; Agarwal, P.; Starczynowski, D.T. TIFA and TIFAB: FHA-domain proteins involved in inflammation, hematopoiesis, and disease. Exp. Hematol. 2020, 90, 18–29. [Google Scholar] [CrossRef]
- Zhang, C.-F.; Wang, H.-M.; Wu, A.; Li, Y.; Tian, X.-L. FHA domain of AGGF1 is essential for its nucleocytoplasmic transport and angiogenesis. Sci. China Life Sci. 2021, 1–11. [Google Scholar] [CrossRef]
- Gelb, M.H.; Cho, W.; Wilton, D.C. Interfacial binding of secreted phospholipases A2: More than electrostatics and a major role for tryptophan. Curr. Opin. Struct. Biol. 1999, 9, 428–432. [Google Scholar] [CrossRef]
- Watanabe, L.; Gava, L.M.; Angulo, Y.; Lomonte, B.; Arni, R.K. Crystallization of the Lys49 PLA2 homologue, myotoxin II, from the venom of Atropoides nummifer. Biochim. Biophys. Acta 2004, 1703, 87–89. [Google Scholar] [CrossRef] [PubMed]
- Delatorre, P.; Rocha, B.A.M.; Santi-Gadelha, T.; Gadelha, C.A.A.; Toyama, M.H.; Cavada, B.S. Crystal structure of Bn IV in complex with myristic acid: A Lys49 myotoxic phospholipase A₂ from Bothrops neuwiedi venom. Biochimie 2011, 93, 513–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakane, F.; Hoshino, F.; Murakami, C. New Era of Diacylglycerol Kinase, Phosphatidic Acid and Phosphatidic Acid-Binding Protein. Int. J. Mol. Sci. 2020, 21, 6794. [Google Scholar] [CrossRef]
- Mahalka, A.K.; Code, C.; Jahromi, B.R.; Kirkegaard, T.; Jäättelä, M.; Kinnunen, P.K. Activation of phospholipase A2 by Hsp70 in vitro. Biochim. Biophys. Acta 2011, 1808, 2569–2572. [Google Scholar] [CrossRef] [Green Version]
- Code, C.; Domanov, Y.; Jutila, A.; Kinnunen, P.K.J. Amyloid-Type Fiber Formation in Control of Enzyme Action: Interfacial Activation of Phospholipase A2. Biophys. J. 2008, 95, 215–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Code, C.; Domanov, Y.A.; Killian, J.A.; Kinnunen, P.K. Activation of phospholipase A2 by temporin B: Formation of antimicrobial peptide-enzyme amyloid-type cofibrils. Biochim. Biophys. Acta 2009, 1788, 1064–1072. [Google Scholar] [CrossRef] [Green Version]
- Cascarina, S.M.; Ross, E.D. Yeast prions and human prion-like proteins: Sequence features and prediction methods. Cell. Mol. Life Sci. 2014, 71, 2047–2063. [Google Scholar] [CrossRef] [Green Version]
- Moreira, V.; de Castro Souto, P.C.M.; Ramirez Vinolo, M.A.; Lomonte, B.; María Gutiérrez, J.; Curi, R.; Teixeira, C. A catalytically-inactive snake venom Lys49 phospholipase A₂ homolog induces expression of cyclooxygenase-2 and production of prostaglandins through selected signaling pathways in macrophages. Eur. J. Pharmacol. 2013, 708, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Dore, E.; Boilard, E. Roles of secreted phospholipase A2 group IIA in inflammation and host defense. Biochim. Biophys. Acta 2019, 1864, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.P.; Leiguez, E.; Guijas, C.; Lomonte, B.; Gutiérrez, J.M.; Teixeira, C.; Balboa, M.A.; Balsinde, J. A Lipidomic Perspective of the Action of Group IIA Secreted Phospholipase A2 on Human Monocytes: Lipid Droplet Biogenesis and Activation of Cytosolic Phospholipase A2α. Biomolecules 2020, 10, 891. [Google Scholar] [CrossRef]
- Ren, S.; Uversky, V.N.; Chen, Z.; Dunker, A.K.; Obradovic, Z. Short Linear Motifs recognized by SH2, SH3 and Ser/Thr Kinase domains are conserved in disordered protein regions. BMC Genom. 2008, 9 (Suppl. S2), S26. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, S.S.; Silveira, L.B.; Da Silva, F.M.N.; Marchi-Salvador, D.P.; Silva, F.P.; Izidoro, L.F.M.; Fuly, A.L.; Juliano, M.A.; Dos Santos, C.R.; Murakami, M.T.; et al. Molecular characterization of an acidic phospholipase A2 from Bothrops pirajai snake venom: Synthetic C-terminal peptide identifies its antiplatelet region. Arch. Toxicol. 2011, 85, 1219–1233. [Google Scholar] [CrossRef] [PubMed]
- Chioato, L.; De Oliveira, A.H.; Ruller, R.; Sá, J.M.; Ward, R.J. Distinct sites for myotoxic and membrane-damaging activities in the C-terminal region of a Lys49-phospholipase A2. Biochem. J. 2002, 366, 971–976. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.I.; Dua, R.; Cho, W. A Structural Determinant of the Unique Interfacial Binding Mode of Bovine Pancreatic Phospholipase A2. Biochemistry 1999, 38, 7811–7818. [Google Scholar] [CrossRef]
- Carman, P.J.; Dominguez, R. BAR domain proteins—a linkage between cellular membranes, signaling pathways, and the actin cytoskeleton. Biophys. Rev. 2018, 10, 1587–1604. [Google Scholar] [CrossRef]
- Suzuki, T.; Bridges, D.; Nakada, D.; Skiniotis, G.; Morrison, S.J.; Lin, J.D.; Saltiel, A.R.; Inoki, K. Inhibition of AMPK Catabolic Action by GSK3. Mol. Cell 2013, 50, 407–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souder, D.C.; Anderson, R.M. An expanding GSK3 network: Implications for aging research. Geroscience 2019, 41, 369–382. [Google Scholar] [CrossRef]
- Nakatsu, Y.; Yamamotoya, T.; Ueda, K.; Ono, H.; Inoue, M.-K.; Matsunaga, Y.; Kushiyama, A.; Sakoda, H.; Fujishiro, M.; Matsubara, A.; et al. Prolyl isomerase Pin1 in metabolic reprogramming of cancer cells. Cancer Lett. 2020, 470, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Liou, Y.-C.; Zhou, X.Z.; Lu, K.P. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem. Sci. 2011, 36, 501–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuefner, M.S.; Pham, K.; Redd, J.R.; Stephenson, E.J.; Harvey, I.; Deng, X.; Bridges, D.; Boilard, E.; Elam, M.B.; Park, E.A. Secretory phospholipase A. J. Lipid Res. 2017, 58, 1822–1833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madej, T.; Lanczycki, C.J.; Zhang, D.; Thiessen, P.A.; Geer, R.C.; Marchler-Bauer, A.; Bryant, S.H. MMDB and VAST+: Tracking structural similarities between macromolecular complexes. Nucleic Acids Res. 2014, 42, D297–D303. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peggion, C.; Tonello, F. Short Linear Motifs Characterizing Snake Venom and Mammalian Phospholipases A2. Toxins 2021, 13, 290. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13040290
Peggion C, Tonello F. Short Linear Motifs Characterizing Snake Venom and Mammalian Phospholipases A2. Toxins. 2021; 13(4):290. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13040290
Chicago/Turabian StylePeggion, Caterina, and Fiorella Tonello. 2021. "Short Linear Motifs Characterizing Snake Venom and Mammalian Phospholipases A2" Toxins 13, no. 4: 290. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13040290