Bothrops moojeni Venom and Its Components Strongly Affect Osteoclasts’ Maturation and Protein Patterns
Abstract
:1. Introduction
2. Results and Discussion
2.1. Effect of B. moojeni Crude Venom on Cell Viability, TRAP+ OCs Number, and F-Acting Ring Integrity
2.2. Effect of Low and High Molecular Mass Fraction of B. moojeni on Viability and TRAP+ OCs Number, and F-Acting Ring Integrity
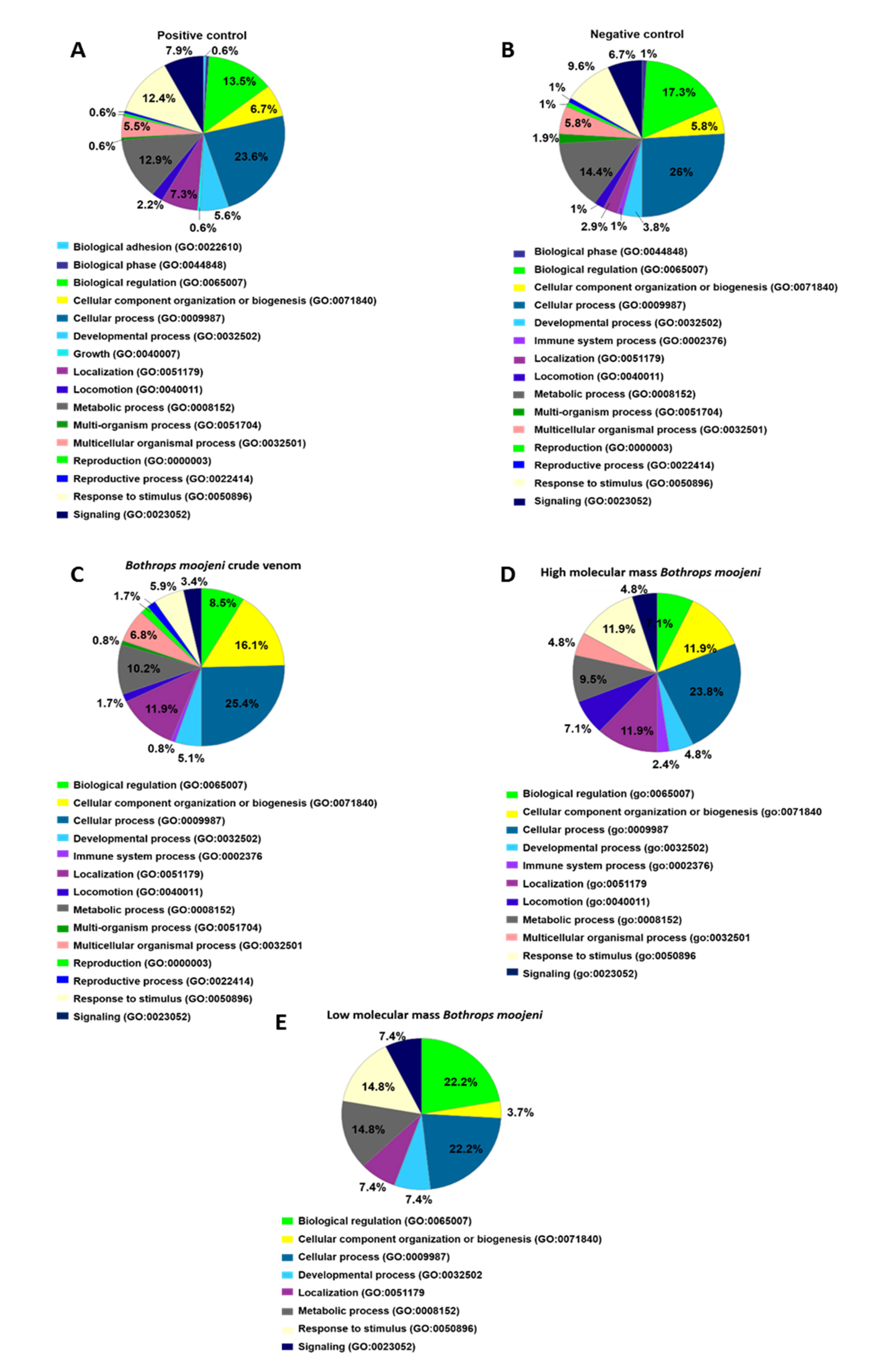
2.3. Cell Culture Medium Soluble Protein Analysis of Mature OCs (Day 15)
2.4. Secreted and Non-Secreted Proteins Identified in the Positive Control of Mature OCs
2.5. Secreted and Non-Secreted Proteins Identified in Mature OCs after Treatment with Crude Venom and Its Components
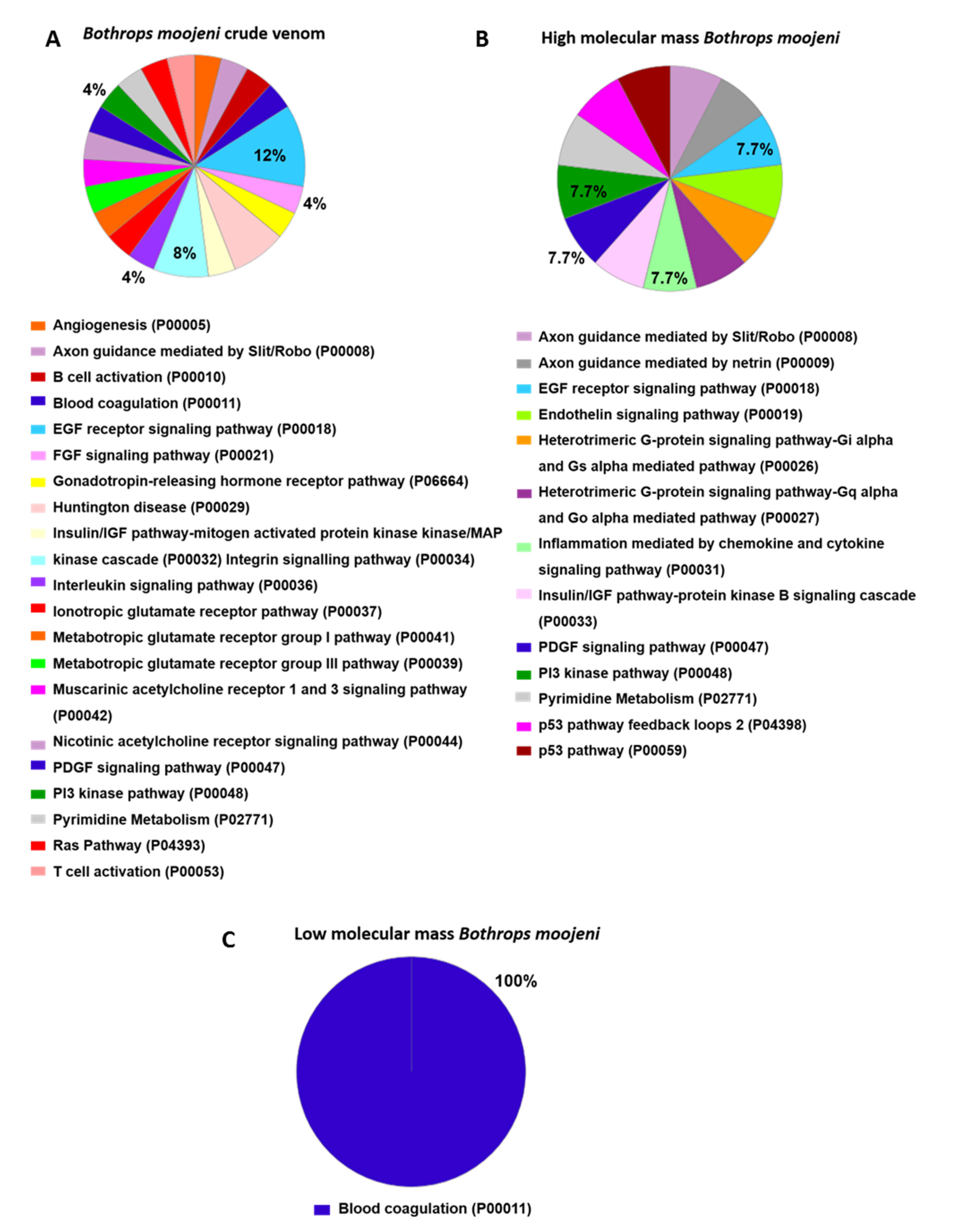
2.6. OCs Signaling Pathways Identified in the Positive Control
2.7. Osteoclastogenesis-Related Signaling Pathways Identified in OCs Treated with Crude Venom or LMM and HMM Fractions
3. Conclusions
4. Materials and Methods
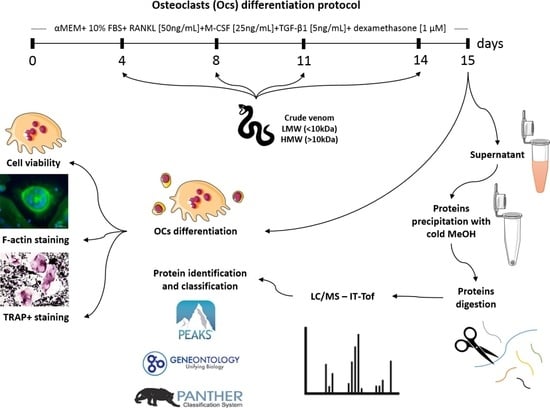
4.1. PBMCs and Osteoclast Differentiation Protocol
4.2. Venom Samples Preparation
4.3. Cell Viability Test—Cell Count Kit 8 (CCK8)
4.4. Phosphatase Resistant of Tartarate (TRAP) Staining
4.5. F-Actin Ring Staining
4.6. Secretome by in Solution Digestion and Mass Spectrometry Analyses
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pinho, F.M.O.; Pereira, I.D. Ofidismo. Rev. Assoc. Médica Bras. 2001, 47, 24–29. [Google Scholar] [CrossRef]
- Amorim, F.G.; Costa, T.R.; Baiwir, D.; De Pauw, E.; Quinton, L.; Sampaio, S.V. Proteopeptidomic, functional and immunoreactivity characterization of Bothrops moojeni snake venom: Influence of snake gender on venom composition. Toxins 2018, 10, 177. [Google Scholar] [CrossRef] [Green Version]
- Mamede, C.C.N.; de Sousa, B.B.; da Cunha Pereira, D.F.; Matias, M.S.; de Queiroz, M.R.; de Morais, N.C.G.; Vieira, S.A.P.B.; Stanziola, L.; de Oliveira, F. Comparative analysis of local effects caused by Bothrops alternatus and Bothrops moojeni snake venoms: Enzymatic contributions and inflammatory modulations. Toxicon 2016, 117, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Weissman, I.L.; Anderson, D.J.; Gage, F. Stem and progenitor cells: Origins, phenotypes, lineage commitments, and transdifferentiations. Annu. Rev. Cell Dev. Biol. 2001, 17, 387–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amarasekara, D.S.; Yun, H.; Kim, S.; Lee, N.; Kim, H.; Rho, J. Regulation of osteoclast differentiation by cytokine networks. Immune Netw. 2018, 18. [Google Scholar] [CrossRef]
- Owen, M. The origin of bone cells. Int. Rev. Cytol. 1970, 28, 213–238. [Google Scholar] [PubMed]
- Burger, E.H.; Van der Meer, J.W.; Nijweide, P.J. Osteoclast formation from mononuclear phagocytes: Role of bone-forming cells. J. Cell Biol. 1984, 99, 1901–1906. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.; Takeshita, S.; Barker, J.E.; Kanagawa, O.; Ross, F.P.; Teitelbaum, S.L. TNF-α induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. J. Clin. Investig. 2000, 106, 1481–1488. [Google Scholar] [CrossRef] [PubMed]
- Susa, M.; Luong-Nguyen, N.-H.; Cappellen, D.; Zamurovic, N.; Gamse, R. Human primary osteoclasts: In vitro generation and applications as pharmacological and clinical assay. J. Transl. Med. 2004, 2, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Bi, H.; Chen, X.; Gao, S.; Yu, X.; Xiao, J.; Zhang, B.; Liu, X.; Dai, M. Key triggers of osteoclast-related diseases and available strategies for targeted therapies: A review. Front. Med. 2017, 4. [Google Scholar] [CrossRef]
- Rodan, G.A.; Martin, T.J. Therapeutic approaches to bone diseases. Science 2000, 289, 1508–1514. [Google Scholar] [CrossRef]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef] [PubMed]
- D’Amélio, F.; Barros, H.; Silva, Á.; Frare, E.; Batista, I.; Pimenta, D.; Kerkis, I. Bothrops moojeni venom: A new tool to investigate osteoclasts differentiation. Sciforum 2021. [Google Scholar] [CrossRef]
- Mercer, B.; Markland, F.; Minkin, C. Contortrostatin, a homodimeric snake venom disintegrin, is a potent inhibitor of osteoclast attachment. J. Bone Miner. Res. 1998, 13, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, I.; Tanaka, H.; Rodan, G.A.; Duong, L.T. Echistatin inhibits the migration of murine prefusion osteoclasts and the formation of multinucleated osteoclast-like cells. Endocrinology 1998, 139, 5182–5193. [Google Scholar] [CrossRef] [PubMed]
- Faloni, A.P.; Sasso-Cerri, E.; Rocha, F.R.; Katchburian, E.; Cerri, P.S. Structural and functional changes in the alveolar bone osteoclasts of estrogen-treated rats. J. Anat. 2012, 220, 77–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longhini, R.; de Oliveira, P.A.; de Souza Faloni, A.P.; Sasso-Cerri, E.; Cerri, P.S. Increased apoptosis in osteoclasts and decreased RANKL immunoexpression in periodontium of cimetidine-treated rats. J. Anat. 2013, 222, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Mambelli-Lisboa, N.C.; Frare, E.O.; da Costa-Neves, A.; Prieto da Silva, Á.R.B.; Kerkis, I. Murine osteoclastogenesis suppression using conditioned media produced by melanoma or activated and non-activated Jurkat-E6 cells. Cell Biol. Int. 2020, 44, 1184–1192. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S.V.; Roodman, G.D. Control of osteoclast differentiation. Crit. Rev. Eukaryot. Gene Exp. 1998, 8, 1–17. [Google Scholar] [CrossRef]
- Biswas, R.S.; Baker, D.A.; Hruska, K.A.; Chellaiah, M.A. Polyphosphoinositides-dependent regulation of the osteoclast actin cytoskeleton and bone resorption. BMC Cell Biol. 2004, 5, 19. [Google Scholar] [CrossRef] [Green Version]
- Choo, M.-J.; Chole, R.A. Cytochrome P-450 inhibition blocks bone resorption in vitro and in vivo. Otolaryngol. Head Neck Surg. 1999, 120, 84–91. [Google Scholar] [CrossRef]
- Zhou, C.; You, Y.; Shen, W.; Zhu, Y.-Z.; Peng, J.; Feng, H.-T.; Wang, Y.; Li, D.; Shao, W.-W.; Li, C.-X. Deficiency of sorting nexin 10 prevents bone erosion in collagen-induced mouse arthritis through promoting NFATc1 degradation. Ann. Rheum. Dis. 2016, 75, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Kohara, Y.; Haraguchi, R.; Kitazawa, R.; Imai, Y.; Kitazawa, S. Hedgehog Inhibitors suppress osteoclastogenesis in in vitro cultures, and deletion of smo in macrophage/osteoclast lineage prevents age-related bone loss. Int. J. Mol. Sci. 2020, 21, 2745. [Google Scholar] [CrossRef] [PubMed]
- Ohba, S. Hedgehog Signaling in Skeletal Development: Roles of Indian Hedgehog and the Mode of Its Action. Int. J. Mol. Sci. 2020, 21, 6665. [Google Scholar] [CrossRef]
- Kim, H.; Takegahara, N.; Matthew, C.; Walsh, J.U.; Fujihara, Y.; Ikawa, M.; Choi, Y. Protocadherin-7 contributes to maintenance of bone homeostasis through regulation of osteoclast multinucleation. BMB Rep. 2020, 53, 472. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Nakashima, T.; Taniguchi, M.; Kodama, T.; Kumanogoh, A.; Takayanagi, H. Osteoprotection by semaphorin 3A. Nature 2012, 485, 69–74. [Google Scholar] [CrossRef]
- Narahara, S.; Sakai, E.; Kadowaki, T.; Yamaguchi, Y.; Narahara, H.; Okamoto, K.; Asahina, I.; Tsukuba, T. KBTBD11, a novel BTB-Kelch protein, is a negative regulator of osteoclastogenesis through controlling Cullin3-mediated ubiquitination of NFATc1. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiang, Y.-W.; Heuck, C.J.; Shaughnessy, J.D., Jr.; Barlogie, B.; Epstein, J. Proteasome inhibitors and bone disease. Semin. Hematol. 2012, 49, 243–248. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, D.N.; Kondo, M.Y.; Oliveira, L.C.G.; Honorato, R.V.; Zanphorlin, L.M.; Coronado, M.A.; Araújo, M.S.; da Motta, G.; Veronez, C.L.; Andrade, S.S. PI class metalloproteinase from Bothrops moojeni venom is a post-proline cleaving peptidase with kininogenase activity: Insights into substrate selectivity and kinetic behavior. Biochim. Biophys. Acta 2014, 1844, 545–552. [Google Scholar] [CrossRef] [Green Version]
- Tiedemann, K.; Boraschi-Diaz, I.; Rajakumar, I.; Kaur, J.; Roughley, P.; Reinhardt, D.P.; Komarova, S.V. Fibrillin-1 directly regulates osteoclast formation and function by a dual mechanism. J. Cell Sci. 2013, 126, 4187–4194. [Google Scholar] [CrossRef] [Green Version]
- Baba, M.; Endoh, M.; Ma, W.; Toyama, H.; Hirayama, A.; Nishikawa, K.; Takubo, K.; Hano, H.; Hasumi, H.; Umemoto, T. Folliculin regulates osteoclastogenesis through metabolic regulation. J. Bone Miner. Res. 2018, 33, 1785–1798. [Google Scholar] [CrossRef] [PubMed]
- Colucci, S.; Colaianni, G.; Mori, G.; Grano, M.; Zallone, A. Human osteoclasts express oxytocin receptor. Biochem. Biophys. Res. Commun. 2002, 297, 442–445. [Google Scholar] [CrossRef]
- Biosse-Duplan, M.; Baroukh, B.; Dy, M.; de Vernejoul, M.-C.; Saffar, J.-L. Histamine promotes osteoclastogenesis through the differential expression of histamine receptors on osteoclasts and osteoblasts. Am. J. Pathol. 2009, 174, 1426–1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brakspear, K.; Mason, D. Glutamate signalling in bone. Front. Endocrinol. 2012, 3, 97. [Google Scholar]
- Espinosa, L.; Itzstein, C.; Cheynel, H.; Delmas, P.D.; Chenu, C. Active NMDA glutamate receptors are expressed by mammalian osteoclasts. J. Physiol. 1999, 518, 47–53. [Google Scholar] [CrossRef]
- Adapala, N.S.; Root, S.; Lorenzo, J.; Aguila, H.; Sanjay, A. PI3K activation increases SDF-1 production and number of osteoclast precursors, and enhances SDF-1-mediated osteoclast precursor migration. Bone Rep. 2019, 10, 100203. [Google Scholar] [CrossRef]
- Hoebertz, A.; Arnett, T.R.; Burnstock, G. Regulation of bone resorption and formation by purines and pyrimidines. Trends Pharmacol. Sci. 2003, 24, 290–297. [Google Scholar] [CrossRef]
- Nascimento, N.G.; Sampaio, M.C.; Olivo, R.A.; Teixeira, C. Contribution of mast cells to the oedema induced by Bothrops moojeni snake venom and a pharmacological assessment of the inflammatory mediators involved. Toxicon 2010, 55, 343–352. [Google Scholar] [CrossRef]
- Nadur-Andrade, N.; Barbosa, A.M.; Carlos, F.P.; Lima, C.J.; Cogo, J.C.; Zamuner, S.R. Effects of photobiostimulation on edema and hemorrhage induced by Bothrops moojeni venom. Lasers Med. Sci. 2012, 27, 65–70. [Google Scholar] [CrossRef]
- Assakura, M.T.; Mandelbaum, F.R. Cleavage of immunoglobulins by moojeni protease A, from the venom of Bothrops moojeni. Toxicon 1990, 28, 734–736. [Google Scholar] [CrossRef]
- Silva, M.C.; Lopes Silva, T.; Silva, M.V.; Mota, C.M.; Santiago, F.M.; Fonseca, K.C.; Oliveira, F.; Mineo, T.W.P.; Mineo, J.R. Interaction between TNF and BmooMP-alpha-I, a zinc metalloprotease derived from Bothrops moojeni snake venom, promotes direct proteolysis of this cytokine: Molecular modeling and docking at a glance. Toxins 2016, 8, 223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kouyoumdjian, J.A.; Polizelli, C. Acidentes ofídicos causados por Bothrops moojeni: Correlação do quadro clínico com o tamanho da serpente. Rev. Inst. Med. Trop. São Paulo 1989, 31, 84–90. [Google Scholar] [CrossRef]
- Kouyoumdjian, J.A.; Polizelli, C. Acidentes ofídicos causados por Bothrops moojeni: Relato de 37 casos. Rev. Inst. Med. Trop. São Paulo 1988, 30, 424–432. [Google Scholar] [CrossRef] [Green Version]
- Boer-Lima, P.A.; Gontijo, J.A.; da Cruz-Höfling, M.A. Histologic and functional renal alterations caused by Bothrops moojeni snake venom in rats. Am. J. Trop. Med. Hyg. 1999, 61, 698–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira-Crott, L.S.; Casare-Ogasawara, T.M.; Ambrosio, L.; Chaim, L.F.P.; de Morais, F.R.; Cintra, A.C.O.; Canicoba, N.C.; Tucci, L.F.F.; Torqueti, M.R.; Sampaio, S.V. Bothrops moojeni venom and BmooLAAO-I downmodulate CXCL8/IL-8 and CCL2/MCP-1 production and oxidative burst response, and upregulate CD11b expression in human neutrophils. Int. Immunopharmacol. 2020, 80, 106154. [Google Scholar] [CrossRef]
- Assakura, M.T.; Reichl, A.P.; Asperti, M.C.A.; Mandelbaum, F.R. Isolation of the major proteolytic enzyme from the venom of the snake Bothrops moojeni (caissaca). Toxicon 1985, 23, 691–706. [Google Scholar] [CrossRef]
- Tempone, A.G.; Andrade, H.F., Jr.; Spencer, P.J.; Lourenço, C.O.; Rogero, J.R.; Nascimento, N. Bothrops moojeni venom kills Leishmania spp. with hydrogen peroxide generated by its L-amino acid oxidase. Biochem. Biophys. Res. Commun. 2001, 280, 620–624. [Google Scholar] [CrossRef]
- Stábeli, R.G.; Sant’Ana, C.D.; Ribeiro, P.H.; Costa, T.R.; Ticli, F.K.; Pires, M.G.; Nomizo, A.; Albuquerque, S.; Malta-Neto, N.R.; Marins, M. Cytotoxic L-amino acid oxidase from Bothrops moojeni: Biochemical and functional characterization. Int. J. Biol. Macromol. 2007, 41, 132–140. [Google Scholar] [CrossRef]
- Matsubara, T.; Kinbara, M.; Maeda, T.; Yoshizawa, M.; Kokabu, S.; Yamamoto, T.T. Regulation of osteoclast differentiation and actin ring formation by the cytolinker protein plectin. Biochem. Biophys. Res. Commun. 2017, 489, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Zuo, J.; Holliday, L.S. Specialized roles for actin in osteoclasts: Unanswered questions and therapeutic opportunities. Biomolecules 2019, 9, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansen, I.D.C.; Vermeer, J.A.F.; Bloemen, V.; Stap, J.; Everts, V. Osteoclast fusion and fission. Calcif. Tissue Int. 2012, 90, 515–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavlos, N.J.; Ng, P.Y. “Fusion and fission” unveils remarkable insights into osteoclast plasticity. Calcif. Tissue Int. 2012, 91, 157. [Google Scholar] [CrossRef] [PubMed]




| Positive Control | |||||
| Accession | Coverage (%) | Peptides | Avg. Mass | Description | Secretory |
| O43323|DHH_HUMAN | 1 | 1 | 43,577 | Desert hedgehog protein | SP |
| Q14623|IHH_HUMAN | 1 | 1 | 45,251 | Indian hedgehog protein | SP |
| Q15465|SHH_HUMAN | 1 | 1 | 49,607 | Sonic hedgehog protein | NS |
| Q8N118|CP4X1_HUMAN | 1 | 1 | 58,875 | Cytochrome P450 4X1 | NS |
| O95025|SEM3D_HUMAN | 1 | 1 | 89,651 | Semaphorin-3D | SP |
| Q96JQ0|PCD16_HUMAN | 0 | 1 | 346,182 | Protocadherin-16 | NS |
| Q96A65|EXOC4_HUMAN | 1 | 1 | 110,498 | Exocyst complex component 4 | NS |
| Q9Y5X2|SNX8_HUMAN | 1 | 1 | 52,569 | Sorting nexin-8 | NS |
| Negative Control | |||||
| Accession | Coverage (%) | Peptides | Avg. Mass | Description | Secretory |
| Q8WUM0|NU133_HUMAN | 0 | 1 | 128,979 | Nuclear pore complex protein Nup133 | NS |
| O00206|TLR4_HUMAN | 1 | 1 | 95,680 | Toll-like receptor 4 | NS |
| Q8N7J2|AMER2_HUMAN | 1 | 1 | 69,507 | APC membrane recruitment protein 2 | NS |
| Bothrops moojeni Crude Venom | |||||
| Accession | Coverage (%) | Peptides | Avg. Mass | Description | Secretory |
| Q8N682|DRAM1_HUMAN | 2 | 1 | 26,253 | DNA damage-regulated autophagy modulator protein 1 | NS |
| Q29RF7|PDS5A_HUMAN | 0 | 1 | 150,830 | Sister chromatid cohesion protein PDS5 homolog A | NS |
| Q8N3X6|LCORL_HUMAN | 1 | 1 | 66,964 | Ligand-dependent nuclear receptor corepressor-like protein | NS |
| P62805|H4_HUMAN | 5 | 1 | 11,367 | Histone H4 | NS |
| P43652|AFAM_HUMAN | 1 | 1 | 69,069 | Afamin | SP |
| Q92599|SEPT8_HUMAN | 1 | 1 | 55,756 | Septin-8 | NS |
| O95150|TNF15_HUMAN | 3 | 1 | 28,087 | Tumor necrosis factor ligand superfamily member 15 | SP |
| Q7Z5K2|WAPL_HUMAN | 1 | 1 | 132,945 | Wings apart-like protein homolog | NS |
| High Molecular Mass of Bothrops moojeni Venom | |||||
| Accession | Coverage (%) | Peptides | Avg. Mass | Description | Secretory |
| P51686|CCR9_HUMAN | 1 | 1 | 42,016 | C-C chemokine receptor type 9 | NS |
| Q2WGJ6|KLH38_HUMAN | 1 | 1 | 65,541 | Kelch-like protein 38 | NS |
| Q8WYR1|PI3R5_HUMAN | 1 | 1 | 97,348 | Phosphoinositide 3-kinase regulatory subunit 5 | NS |
| Q14997|PSME4_HUMAN | 0 | 1 | 211,332 | Proteasome activator complex subunit 4 | NS |
| Low Molecular Mass of Bothrops moojeni Venom | |||||
| Accession | Coverage (%) | Peptides | Avg. Mass | Description | Secretoy |
| P01023|A2MG_HUMAN | 1 | 1 | 163,290 | Alpha-2-macroglobulin | SP |
| P35555|FBN1_HUMAN | 0 | 1 | 312,297 | Fibrillin-1 | SP |
| Q8TF40|FNIP1_HUMAN | 0 | 1 | 130,555 | Folliculin-interacting protein 1 | NS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Amélio, F.; Vigerelli, H.; Prieto-da-Silva, Á.R.d.B.; Osório Frare, E.; Batista, I.d.F.C.; Pimenta, D.C.; Kerkis, I. Bothrops moojeni Venom and Its Components Strongly Affect Osteoclasts’ Maturation and Protein Patterns. Toxins 2021, 13, 459. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13070459
D’Amélio F, Vigerelli H, Prieto-da-Silva ÁRdB, Osório Frare E, Batista IdFC, Pimenta DC, Kerkis I. Bothrops moojeni Venom and Its Components Strongly Affect Osteoclasts’ Maturation and Protein Patterns. Toxins. 2021; 13(7):459. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13070459
Chicago/Turabian StyleD’Amélio, Fernanda, Hugo Vigerelli, Álvaro Rossan de Brandão Prieto-da-Silva, Eduardo Osório Frare, Isabel de Fátima Correia Batista, Daniel Carvalho Pimenta, and Irina Kerkis. 2021. "Bothrops moojeni Venom and Its Components Strongly Affect Osteoclasts’ Maturation and Protein Patterns" Toxins 13, no. 7: 459. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13070459