Characterization of New Allergens from the Venom of the European Paper Wasp Polistes dominula
Abstract
:1. Introduction
2. Results
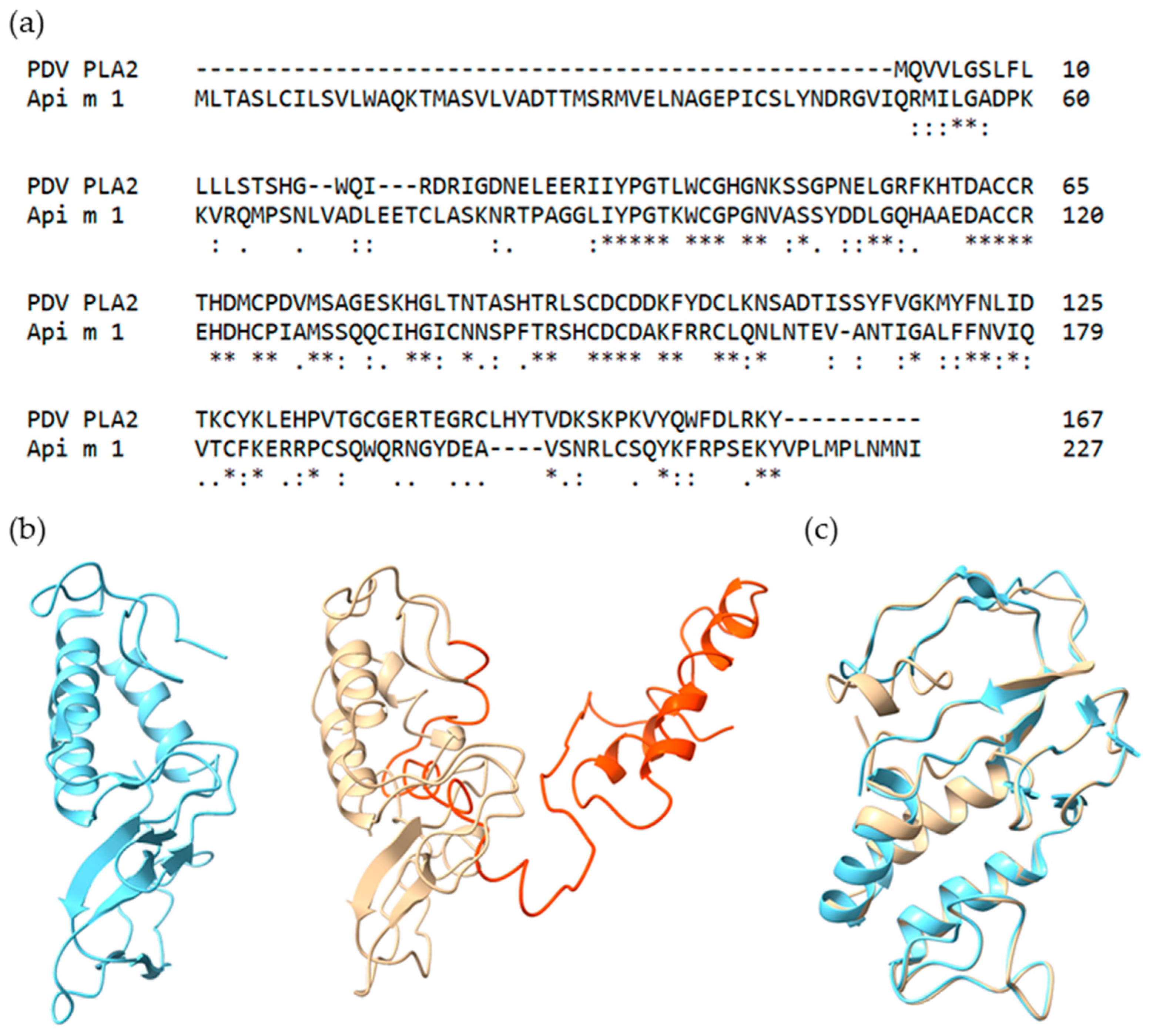
2.1. Selection and Characteristics of Putative PDV Allergens
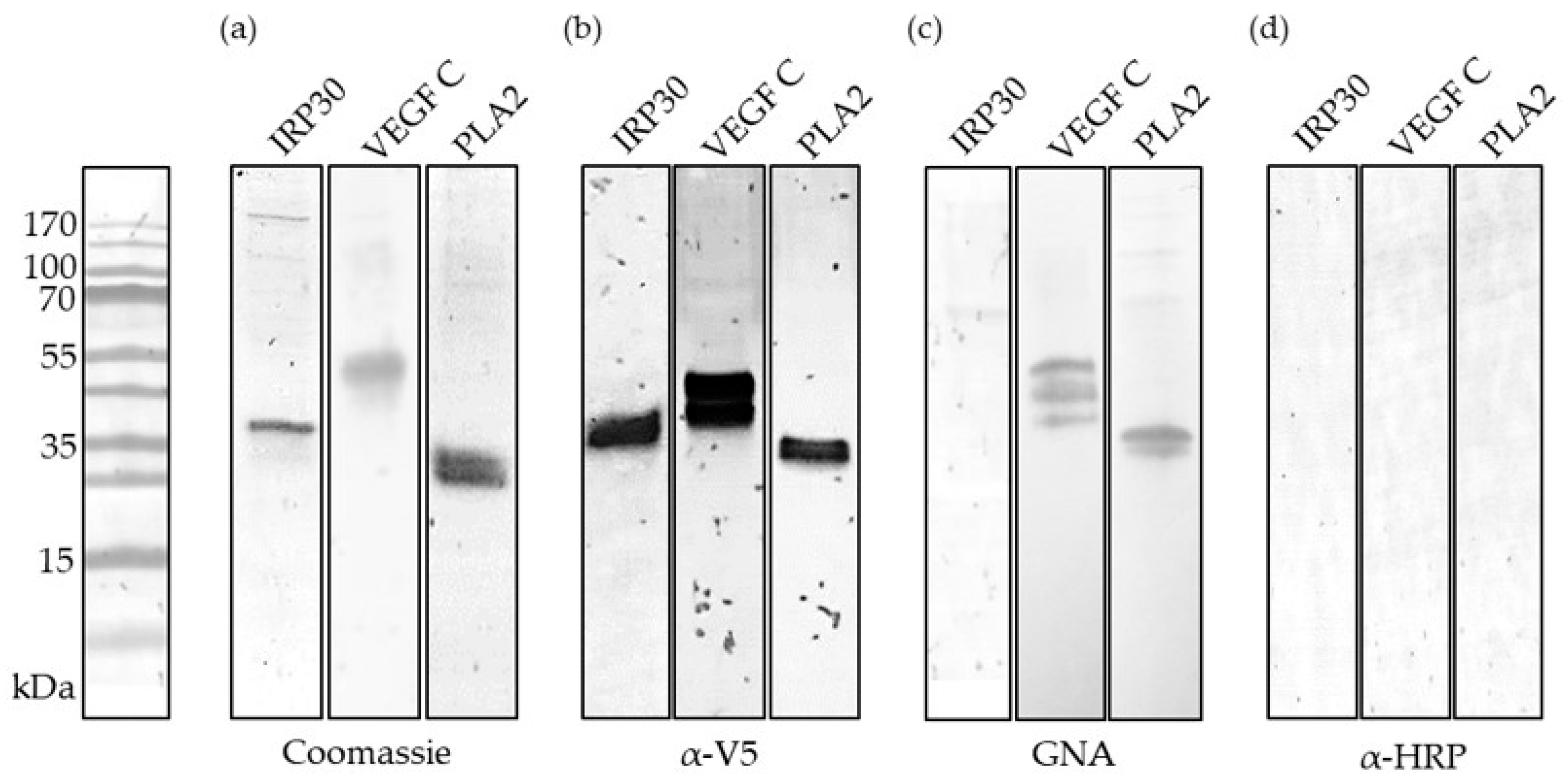
2.2. Recombinant Production and Biochemical Characterization of Putative PDV Allergens
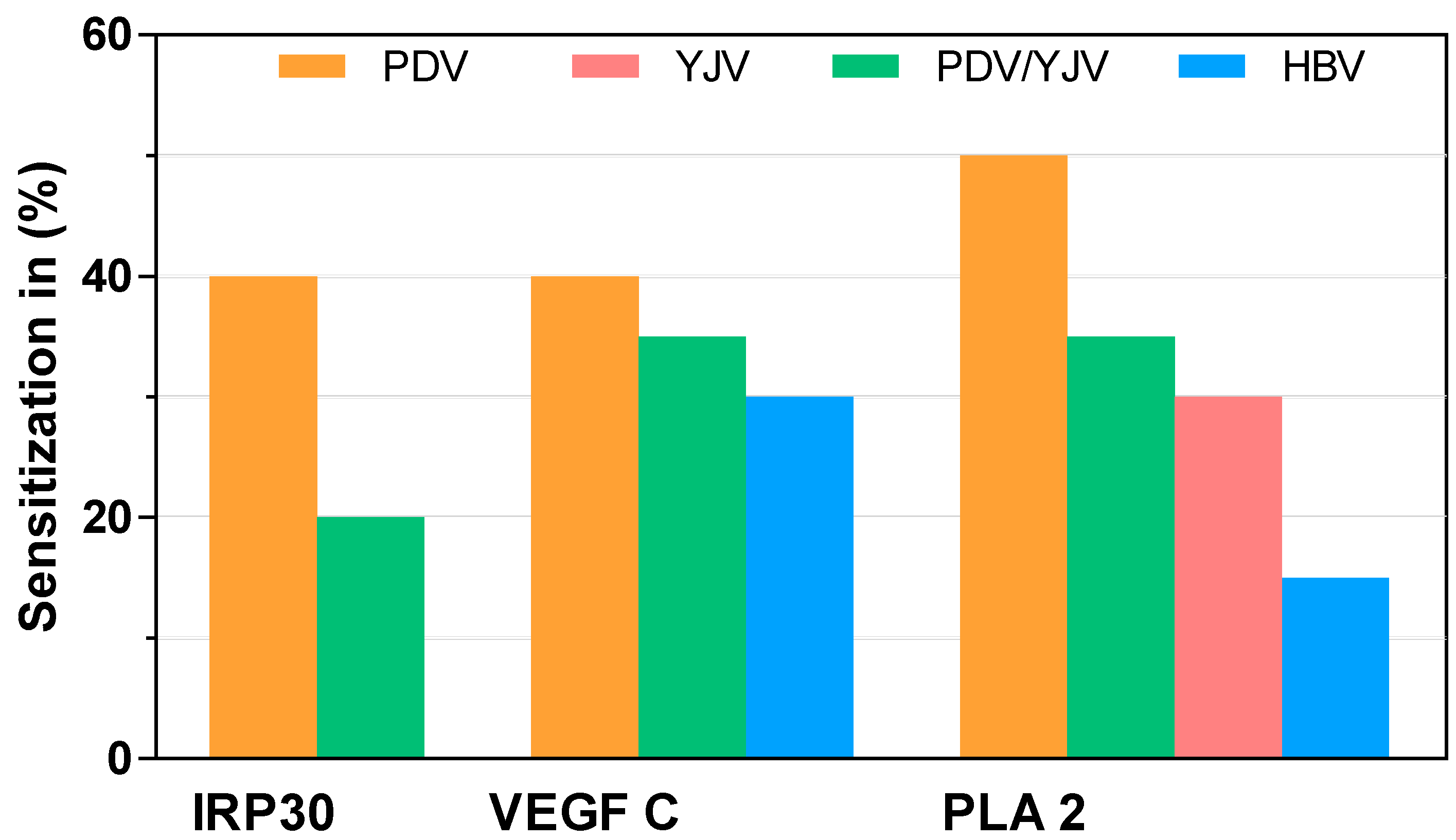
2.3. sIgE Sensitization to Putative PDV Allergens
2.4. Activation of Basophils from Allergic Patients by PDV PLA2 and HBV Api m 1
3. Discussion
4. Materials and Methods
4.1. In silico Tools
4.2. Cloning
4.3. Generation of Recombinant Baculovirus and Recombinant Protein Production
4.4. SDS-PAGE and Western Blotting
4.5. Patients
4.6. sIgE Reactivity of Patient Sera
4.7. Basophil Activation Test
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cervo, R.; Zacchi, F.; Turillazzi, S. Polistes dominulus (Hymenoptera, Vespidae) invading North America: Some hypotheses for its rapid spread. Insectes Soc. 2000, 47, 155–157. [Google Scholar] [CrossRef]
- Eardley, C.; Koch, F.; Wood, A.R. Polistes dominulus (Christ, 1791) (Hymenoptera: Polistinae: Vespidae) newly recorded from South Africa. Afr. Entomol. 2009, 17, 226–227. [Google Scholar] [CrossRef]
- Duty, I. Notes on the occurrence of Vespidae (Hym.) in Mecklenburg, mainly around Rostock. (Zum Vorkommen von Faltenwespen (Hym., Vespidae) in Mecklenburg mit Schwerpunkt im Raum Rostock.). Entomol. Nachr. Ber. 1997, 41, 113–119. [Google Scholar]
- Kowalczyk, J.K.; Szczepko, K. Remarks on the taxonomy and distribution of two species of paper wasps—Polistes gallicus (Linnaeus, 1767) and P. dominulus (Christ, 1791) (Hymenoptera: Vespidae) in Poland. Wiadomości Entomol. 2003, 22, 69–72. [Google Scholar]
- Rusina, L.Y. Self-organization of populations of polistine wasps (Hymenoptera, Vespidae, Polistinae). Entomol. Rev. 2010, 90, 811–829. [Google Scholar] [CrossRef]
- Demain, J.G.; Gessner, B.D.; McLaughlin, J.B.; Sikes, D.S.; Foote, J.T. Increasing insect reactions in Alaska: Is this related to changing climate? Allergy Asthma Proc. 2009, 30, 238–243. [Google Scholar] [CrossRef] [Green Version]
- Oswalt, M.L.; Foote, J.T.; Kemp, S.F. Anaphylaxis: Report of Two Fatal Yellow Jacket (YJ) Stings in Alaska (AK). J. Allergy Clin. Immunol. 2007, 119, 34. [Google Scholar] [CrossRef]
- Baker, T.W.; Forester, J.P.; Johnson, M.L.; Sikora, J.M.; Stolfi, A.; Stahl, M.C. Stinging insect identification: Are the allergy specialists any better than their patients? Ann. Allergy Asthma Immunol. 2016, 116, 431–434. [Google Scholar] [CrossRef] [PubMed]
- Baker, T.W.; Forester, J.P.; Johnson, M.L.; Stolfi, A.; Stahl, M.C. The HIT study: Hymenoptera Identification Test—How accurate are people at identifying stinging insects? Ann. Allergy Asthma Immunol. 2014, 113, 267–270. [Google Scholar] [CrossRef]
- Blank, S.; Bilò, M.B.; Grosch, J.; Schmidt-Weber, C.B.; Ollert, M.; Jakob, T. Marker allergens in Hymenoptera venom allergy—Characteristics and potential use in precision medicine. Allergo J. Int. 2021, 30, 26–38. [Google Scholar] [CrossRef]
- Grosch, J.; Hilger, C.; Bilò, M.B.; Kler, S.; Schiener, M.; Dittmar, G.; Bernardin, F.; Lesur, A.; Ollert, M.; Schmidt-Weber, C.B.; et al. Shedding light on the venom proteomes of the allergy-relevant hymenoptera Polistes dominula (European paper wasp) and Vespula spp. (Yellow Jacket). Toxins 2020, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Monsalve, R.I.; Vega, A.; Marqués, L.; Miranda, A.; Fernández, J.; Soriano, V.; Cruz, S.; Dominguez-Noche, C.; Sanchez-Morillas, L.; Armisen-Gil, M.; et al. Component-resolved diagnosis of vespid venom-allergic individuals: Phospholipases and antigen 5s are necessary to identify Vespula or Polistes sensitization. Allergy Eur. J. Allergy Clin. Immunol. 2012, 67, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Schiener, M.; Hilger, C.; Eberlein, B.; Pascal, M.; Kuehn, A.; Revets, D.; Planchon, S.; Pietsch, G.; Serrano, P.; Darsow, U.; et al. The high molecular weight dipeptidyl peptidase IV Pol d 3 is a major allergen of Polistes dominula venom. Sci. Rep. 2018, 8, 1318. [Google Scholar] [CrossRef] [PubMed]
- Bilò, M.B.; Martini, M.; Bonadonna, P.; Cinti, B.; Da Re, M.; Gabrielli, O.; Olivieri, F.; Salgarolo, V.; Zanoni, G.; Villalta, D. Prevalence of Pol d 1 sensitization in Polistes dominula allergy and its diagnostic role in vespid double-positivity. J. Allergy Clin. Immunol. Pract. 2021. [Google Scholar] [CrossRef]
- Savi, E.; Peveri, S.; Makri, E.; Pravettoni, V.; Incorvaia, C. Comparing the ability of molecular diagnosis and CAP-inhibition in identifying the really causative venom in patients with positive tests to Vespula and Polistes species. Clin. Mol. Allergy 2016, 14, 4–9. [Google Scholar] [CrossRef] [Green Version]
- Quercia, O.; Cova, V.; Martini, M.; Cortellini, G.; Murzilli, F.; Bignardi, D.; Cilia, M.; Scarpa, A.; Bilo, M.B. CAP-Inhibition, molecular diagnostics, and Total IgE in the evaluation of polistes and vespula double sensitization. Int. Arch. Allergy Immunol. 2018, 177, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Wu, J.; Han, L.; Huang, J.; Wang, D. Novel Characteristics of Immune Responsive Protein IRP30 in the Bumble Bee Bombus lantschouensis (Hymenoptera: Apidae). J. Insect Sci. 2020, 20, 11. [Google Scholar] [CrossRef]
- Albert, Š.; Gätschenberger, H.; Azzami, K.; Gimple, O.; Grimmer, G.; Sumner, S.; Fujiyuki, T.; Tautz, J.; Mueller, M.J. Evidence of a novel immune responsive protein in the Hymenoptera. Insect Biochem. Mol. Biol. 2011, 41, 968–981. [Google Scholar] [CrossRef]
- Patnaik, B.B.; Park, S.Y.; Kang, S.W.; Hwang, H.J.; Wang, T.H.; Park, E.B.; Chung, J.M.; Song, D.K.; Kim, C.; Kim, S.; et al. Transcriptome Profile of the Asian Giant Hornet (Vespa mandarinia) Using Illumina HiSeq 4000 Sequencing: De Novo Assembly, Functional Annotation, and Discovery of SSR Markers. Int. J. Genom. 2016, 2016, 4169587. [Google Scholar] [CrossRef] [Green Version]
- Santos, L.D.; Santos, K.S.; de Souza, B.M.; Arcuri, H.A.; Cunha-Neto, E.; Castro, F.M.; Kalil, J.E.; Palma, M.S. Purification, sequencing and structural characterization of the phospholipase A1from the venom of the social wasp Polybia paulista (Hymenoptera, Vespidae). Toxicon 2007, 50, 923–937. [Google Scholar] [CrossRef]
- Yang, H.; Xu, X.; Ma, D.; Zhang, K.; Lai, R. A phospholipase A1 platelet activator from the wasp venom of Vespa magnifica (Smith). Toxicon 2008, 51, 289–296. [Google Scholar] [CrossRef]
- Atakuziev, B.U.; Nuritova, F.; Usmanov, P.B. Phospholipase A2 from the venom of the spider Eresus niger. Chem. Nat. Compd. 1991, 27, 487–489. [Google Scholar] [CrossRef]
- Harris, J.B.; Scott-Davey, T. Secreted phospholipases A2 of snake venoms: Effects on the peripheral neuromuscular system with comments on the role of phospholipases A2 in disorders of the CNS and their uses in industry. Toxins 2013, 5, 2533–2571. [Google Scholar] [CrossRef] [Green Version]
- Swiontek, K.; Planchon, S.; Ollert, M.; Eyer, F.; Fischer, J.; Hilger, C. Phospholipase A2 Triggers Anaphylaxis to Snake Venom by Repeated Skin Sensitization—A Case Report. J. Investig. Allergol. Clin. Immunol. 2020, 31, 175–177. [Google Scholar] [CrossRef]
- Joukov, V.; Kaipainen, A.; Jeltsch, M.; Pajusola, K.; Olofsson, B.; Kumar, V.; Eriksson, U.; Alitalo, K. Vascular Endothelial Growth Factors VEGF-B and VEGF-C. J. Cell. Physiol. 1997, 173, 211–215. [Google Scholar] [CrossRef]
- Anisimov, A.; Alitalo, A.; Korpisalo, P.; Soronen, J.; Kaijalainen, S.; Leppänen, V.M.; Jeltsch, M.; Yla-Herttuala, S.; Alitalo, K. Activated forms of VEGF-C and VEGF-D provide improved vascular function in skeletal muscle. Circ. Res. 2009, 104, 1302–1312. [Google Scholar] [CrossRef]
- Jha, S.K.; Rauniyar, K.; Karpanen, T.; Leppänen, V.M.; Brouillard, P.; Vikkula, M.; Alitalo, K.; Jeltsch, M. Efficient activation of the lymphangiogenic growth factor VEGF-C requires the C-terminal domain of VEGF-C and the N-terminal domain of CCBE1. Sci. Rep. 2017, 7, 4916. [Google Scholar] [CrossRef] [PubMed]
- Joukov, V.; Sorsa, T.; Kumar, V.; Jeltsch, M.; Claesson-Welsh, L.; Cao, Y.; Saksela, O.; Kalkkinen, N.; Alitalo, K. Proteolytic processing regulates receptor specificity and activity of VEGF-C. EMBO J. 1997, 16, 3898–3911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barreto, G.; Sandelin, J.; Salem, A.; Nordström, D.C.; Waris, E. Toll-like receptors and their soluble forms differ in the knee and thumb basal osteoarthritic joints. Acta Orthop. 2017, 88, 326–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, M.J.; Morandi, E.; Tanasescu, R.; Frakich, N.; Caldano, M.; Onion, D.; Faraj, T.A.; Erridge, C.; Gran, B. The soluble form of toll-like receptor 2 is elevated in serum of multiple sclerosis patients: A novel potential disease biomarker. Front. Immunol. 2018, 9, 457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeBouder, E.; Rey-Nores, J.E.; Rushmere, N.K.; Grigorov, M.; Lawn, S.D.; Affolter, M.; Griffin, G.E.; Ferrara, P.; Schiffrin, E.J.; Morgan, B.P.; et al. Soluble Forms of Toll-Like Receptor (TLR)2 Capable of Modulating TLR2 Signaling Are Present in Human Plasma and Breast Milk. J. Immunol. 2003, 171, 6680–6689. [Google Scholar] [CrossRef] [Green Version]
- Binder, M.; Fierlbeck, G.; King, T.P.; Valent, P.; Bühring, H.J. Individual hymenoptera venom compounds induce upregulation of the basophil activation marker ectonucleotide pyrophosphatase/phosphodiesterase 3 (CD203c) in sensitized patients. Int. Arch. Allergy Immunol. 2002, 129, 160–168. [Google Scholar] [CrossRef] [PubMed]
- White, S.P.; Scott, D.L.; Otwinowski, Z.; Gelb, M.H.; Sigler, P.B. Crystal structure of bee-venom phospholipase A2 in a complex with a transition-state analogue. Science 1990, 250, 1563–1566. [Google Scholar] [CrossRef] [PubMed]
- Russkamp, D.; Van Vaerenbergh, M.; Etzold, S.; Eberlein, B.; Darsow, U.; Schiener, M.; De Smet, L.; Absmaier, M.; Biedermann, T.; Spillner, E.; et al. Characterization of the honeybee venom proteins C1q-like protein and PVF1 and their allergenic potential. Toxicon 2018, 150, 198–206. [Google Scholar] [CrossRef] [Green Version]
- Seismann, H.; Blank, S.; Braren, I.; Greunke, K.; Cifuentes, L.; Grunwald, T.; Bredehorst, R.; Ollert, M.; Spillner, E. Dissecting cross-reactivity in hymenoptera venom allergy by circumvention of α-1,3-core fucosylation. Mol. Immunol. 2010, 47, 799–808. [Google Scholar] [CrossRef]
- Blank, S.; Michel, Y.; Seismann, H.; Plum, M.; Greunke, K.; Grunwald, T.; Bredehorst, R.; Ollert, M.; Braren, I.; Spillner, E. Evaluation of Different Glycoforms of Honeybee Venom Major Allergen Phospholipase A2 (Api m 1) Produced in Insect Cells. Protein Pept. Lett. 2011, 18, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Blank, S.; Seismann, H.; Michel, Y.; McIntyre, M.; Cifuentes, L.; Braren, I.; Grunwald, T.; Darsow, U.; Ring, J.; Bredehorst, R.; et al. Api m 10, a genuine A. mellifera venom allergen, is clinically relevant but underrepresented in therapeutic extracts. Allergy Eur. J. Allergy Clin. Immunol. 2011, 66, 1322–1329. [Google Scholar] [CrossRef]
- Köhler, J.; Blank, S.; Müller, S.; Bantleon, F.; Frick, M.; Huss-Marp, J.; Lidholm, J.; Spillner, E.; Jakob, T. Component resolution reveals additional major allergens in patients with honeybee venom allergy. J. Allergy Clin. Immunol. 2014, 133, 1383–1389.e6. [Google Scholar] [CrossRef] [Green Version]
- Van Vaerenbergh, M.; Debyser, G.; Devreese, B.; de Graaf, D.C. Exploring the hidden honeybee (Apis mellifera) venom proteome by integrating a combinatorial peptide ligand library approach with FTMS. J. Proteom. 2014, 99, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.F.; Alpan, O.; Hoffmann, H.J. Basophil activation test: Mechanisms and considerations for use in clinical trials and clinical practice. Allergy Eur. J. Allergy Clin. Immunol. 2021, 76, 2420–2432. [Google Scholar] [CrossRef]
- Eberlein, B. Basophil Activation as Marker of Clinically Relevant Allergy and Therapy Outcome. Front. Immunol. 2020, 11, 1815. [Google Scholar] [CrossRef] [PubMed]
- Blank, S.; Haemmerle, S.; Jaeger, T.; Russkamp, D.; Ring, J.; Schmidt-Weber, C.B.; Ollert, M. Prevalence of Hymenoptera venom allergy and sensitization in the population-representative German KORA cohort. Allergo J. Int. 2019, 28, 183–191. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauhan, J.S.; Rao, A.; Raghava, G.P.S. In silico Platform for Prediction of N-, O- and C-Glycosites in Eukaryotic Protein Sequences. PLoS ONE 2013, 8, e67008. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Chang, H.Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2016, 10, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Blank, S.; Seismann, H.; Bockisch, B.; Braren, I.; Cifuentes, L.; McIntyre, M.; Ruhl, D.; Ring, J.; Bredehorst, R.; Ollert, M.W.; et al. Identification, Recombinant Expression, and Characterization of the 100 kDa High Molecular Weight Hymenoptera Venom Allergens Api m 5 and Ves v3. J. Immunol. 2010, 184, 5403–5413. [Google Scholar] [CrossRef] [Green Version]
- Schiener, M.; Eberlein, B.; Moreno-Aguilar, C.; Pietsch, G.; Serrano, P.; McIntyre, M.; Schwarze, L.; Russkamp, D.; Biedermann, T.; Spillner, E.; et al. Application of recombinant antigen 5 allergens from seven allergy-relevant Hymenoptera species in diagnostics. Allergy 2017, 72, 98–108. [Google Scholar] [CrossRef]
- Classen, D.C.; Morningstar, J.M.; Shanley, J.D. Detection of antibody to murine cytomegalovirus by enzyme-linked immunosorbent and indirect immunofluorescence assays. J. Clin. Microbiol. 1987, 25, 600–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michel, Y.; McIntyre, M.; Ginglinger, H.; Ollert, M.; Cifuentes, L.; Blank, S.; Spillner, E. The putative serine protease inhibitor Api m 6 from Apis Mellifera venom: Recombinant and structural evaluation. J. Investig. Allergol. Clin. Immunol. 2012, 22, 476–484. [Google Scholar] [PubMed]
- Eberlein, B.; Krischan, L.; Darsow, U.; Ollert, M.; Ring, J. Double positivity to bee and wasp venom: Improved diagnostic procedure by recombinant allergen-based IgE testing and basophil activation test including data about cross-reactive carbohydrate determinants. J. Allergy Clin. Immunol. 2012, 130, 155–161. [Google Scholar] [CrossRef] [PubMed]

| nApi m 1 | PDV PLA2 | |
|---|---|---|
| All patients (30) | 12 | 1 |
| MUC patients (18) | 9 | 1 |
| BCN patients (12) | 3 | - |
| HBV sensitized (15) | 12 | 1 |
| PDV sensitized (7) | 1 | - |
| YJV sensitized (24) | 7 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grosch, J.; Eberlein, B.; Waldherr, S.; Pascal, M.; San Bartolomé, C.; De La Roca Pinzón, F.; Dittmar, M.; Hilger, C.; Ollert, M.; Biedermann, T.; et al. Characterization of New Allergens from the Venom of the European Paper Wasp Polistes dominula. Toxins 2021, 13, 559. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13080559
Grosch J, Eberlein B, Waldherr S, Pascal M, San Bartolomé C, De La Roca Pinzón F, Dittmar M, Hilger C, Ollert M, Biedermann T, et al. Characterization of New Allergens from the Venom of the European Paper Wasp Polistes dominula. Toxins. 2021; 13(8):559. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13080559
Chicago/Turabian StyleGrosch, Johannes, Bernadette Eberlein, Sebastian Waldherr, Mariona Pascal, Clara San Bartolomé, Federico De La Roca Pinzón, Michael Dittmar, Christiane Hilger, Markus Ollert, Tilo Biedermann, and et al. 2021. "Characterization of New Allergens from the Venom of the European Paper Wasp Polistes dominula" Toxins 13, no. 8: 559. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13080559




