Expression Analyses of Genes Related to Multixenobiotic Resistance in Mytilus galloprovincialis after Exposure to Okadaic Acid-Producing Dinophysis acuminata
Abstract
:1. Introduction
2. Results
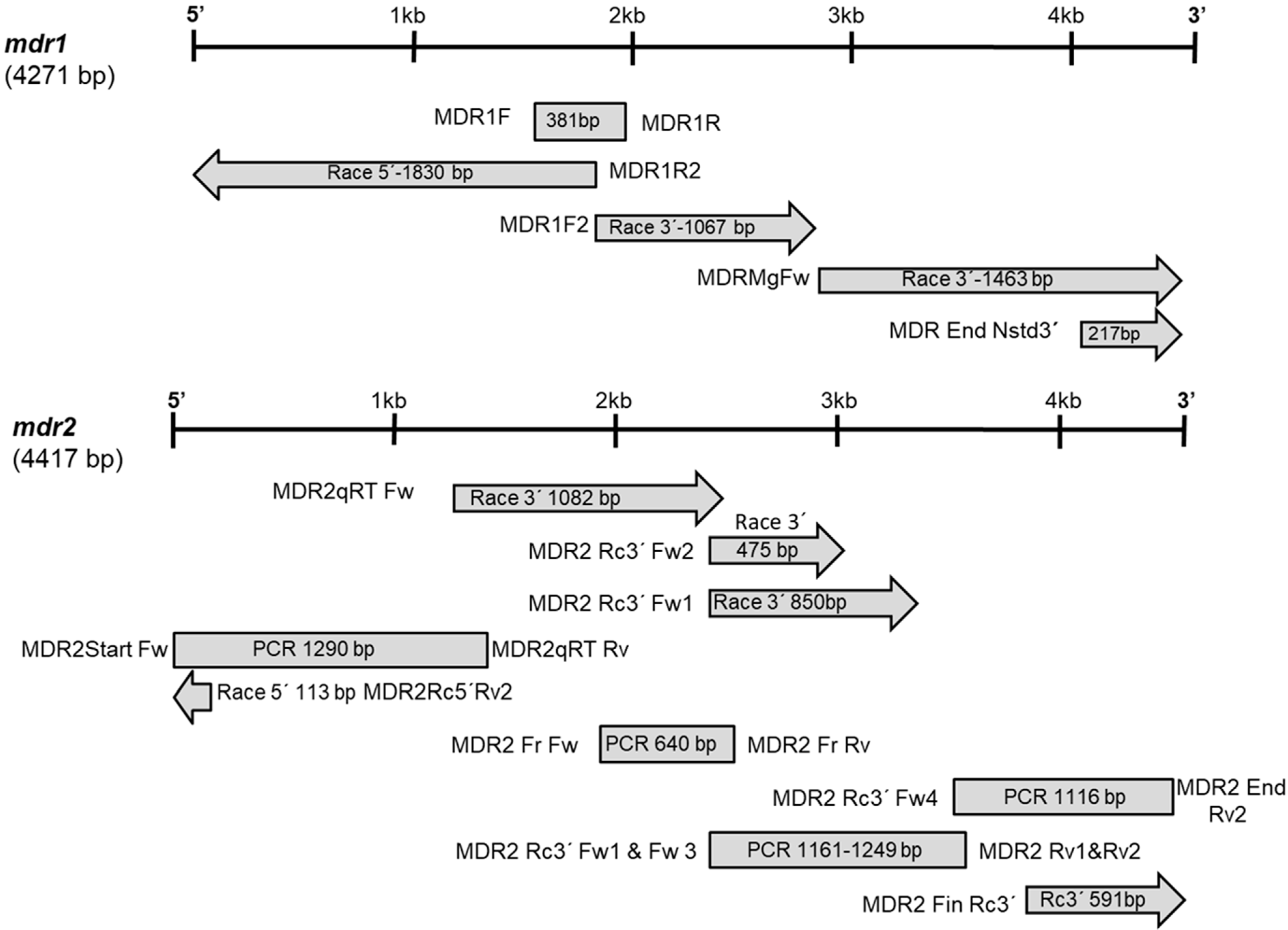
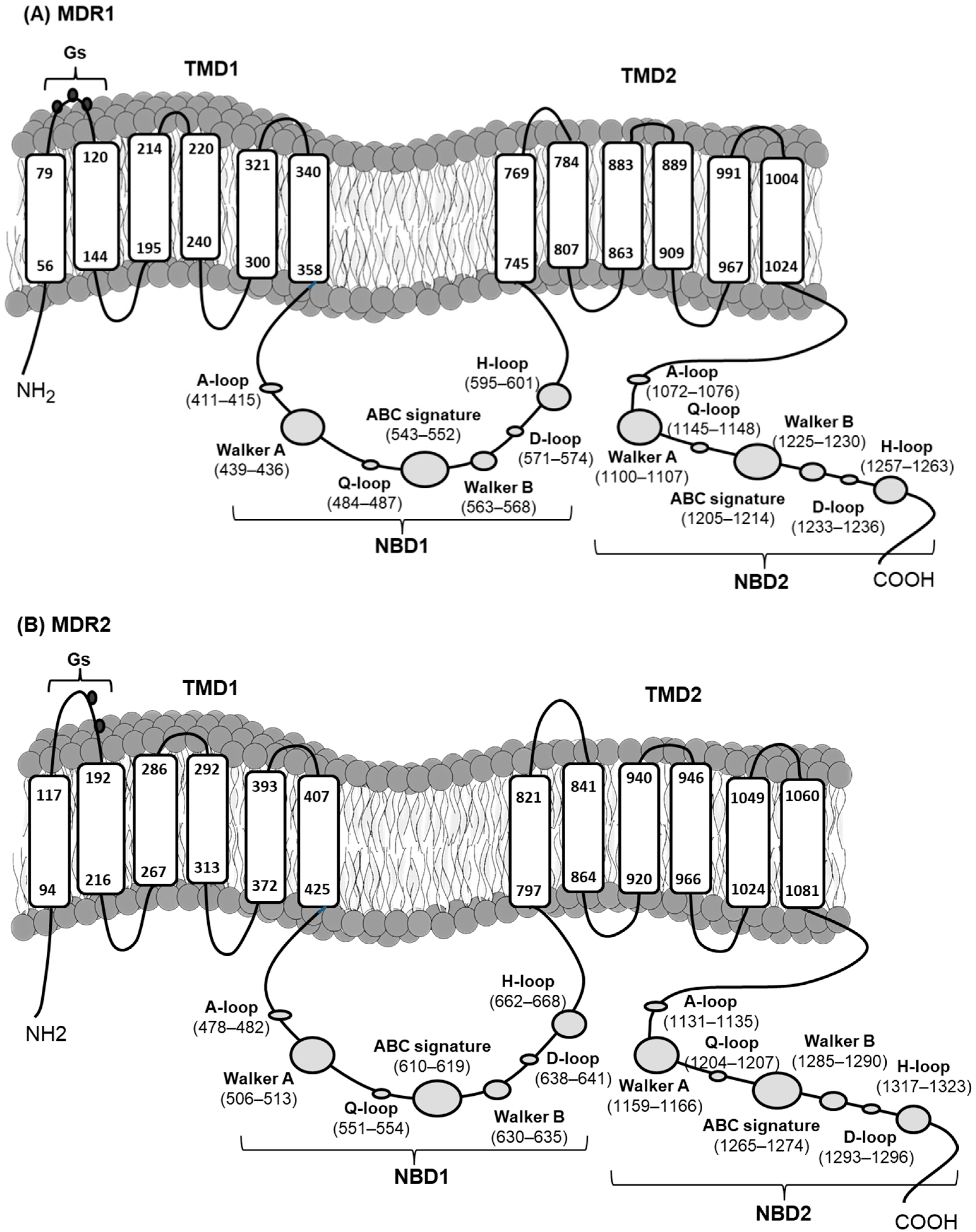
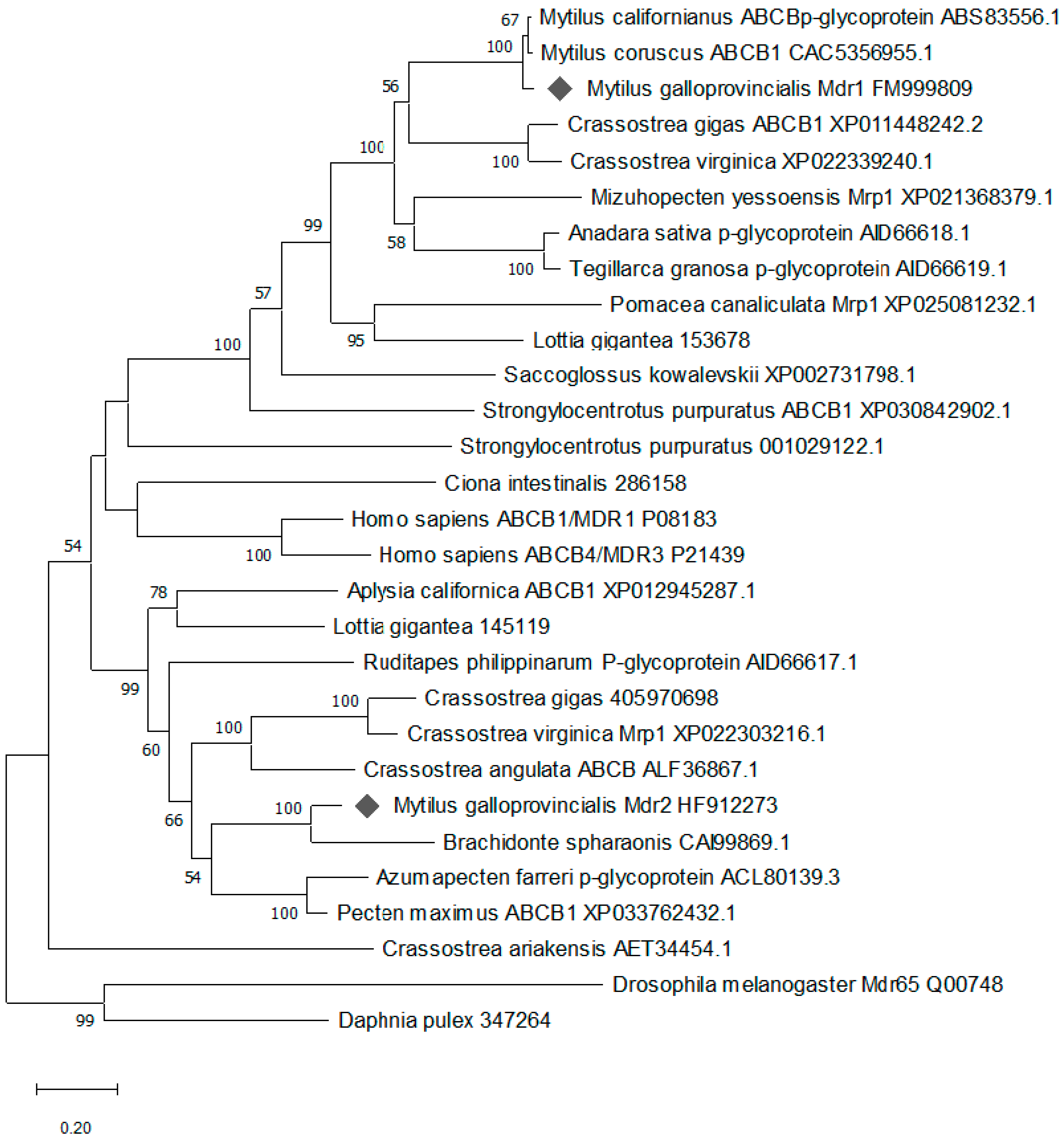
2.1. Cloning and Characterization of mdr Genes

2.2. Gene Expression by RT-qPCR
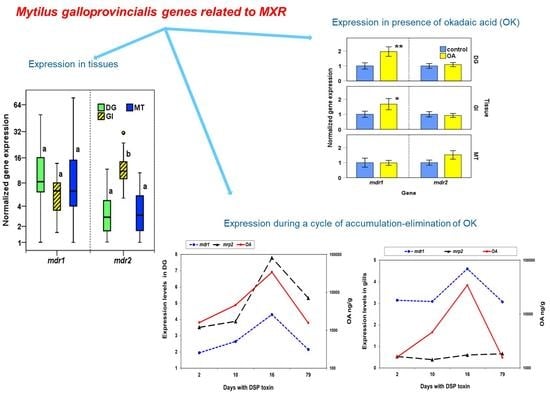
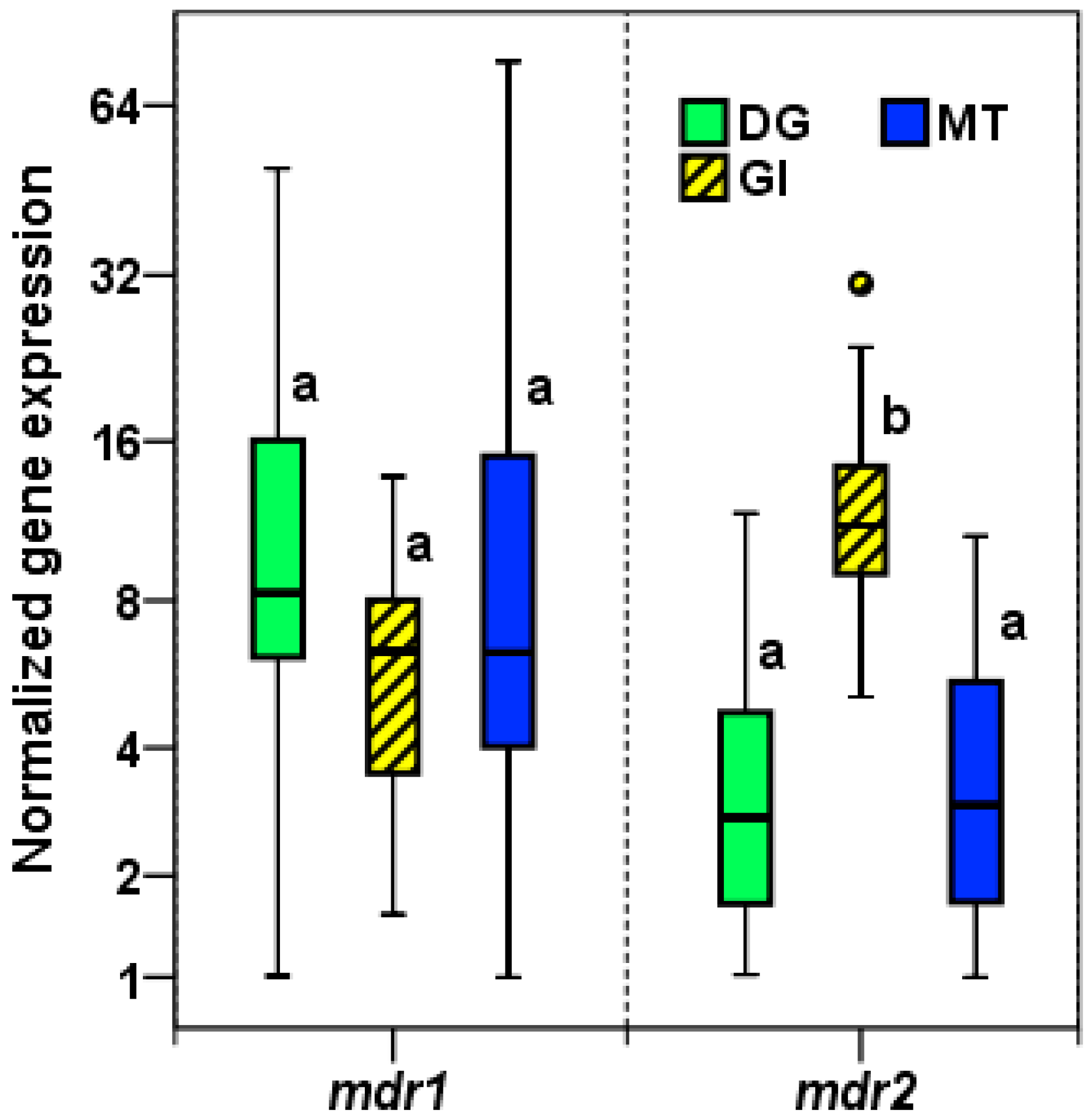
2.2.1. Expression of mdr1 and mdr2 in M. galloprovincialis Tissues
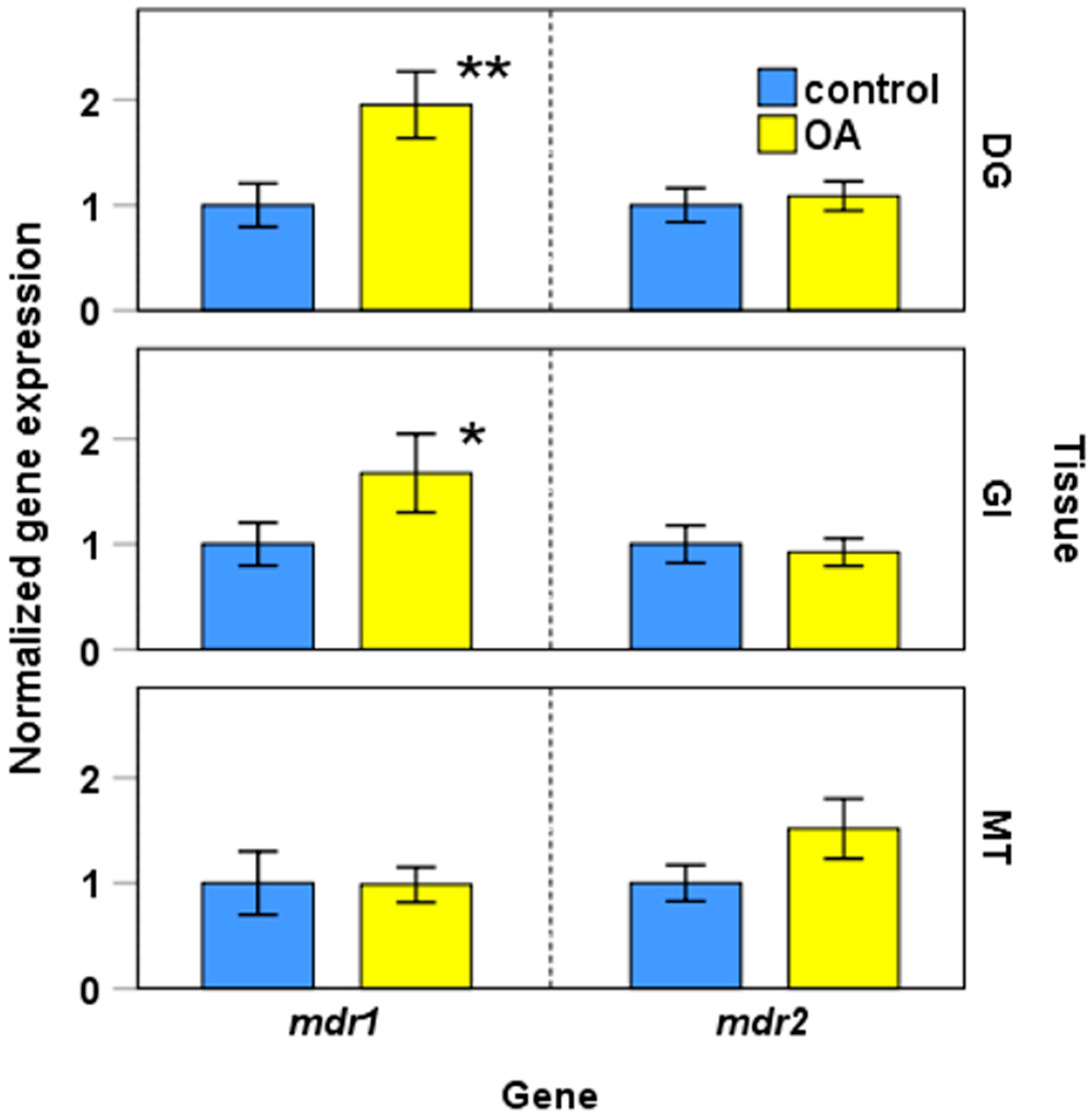
2.2.2. Expression of mdr1, mdr2, and mrp2 in Presence of Okadaic Acid
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cloning and Characterization of mdr Genes
5.1.1. Animals and RNA Extraction
5.1.2. Primer Selection
5.1.3. Cloning of cDNA Fragments and Rapid Amplification of cDNA Ends (RACE)
5.1.4. Sequence and Phylogenetic Analyses
5.2. Expression of mdr and mrp by RT-qPCR
5.2.1. Animals, RNA Extraction, and cDNA Synthesis
5.2.2. Primer Design and PCR Efficiency
5.2.3. Quantitative Real-Time RT-PCR and Data Analysis
5.2.4. Analysis of Gene Expression Stability
5.2.5. Gene Expression and Statistical Analysis
5.3. Toxin Extraction and Okadaic Acid Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moroño, A.; Arévalo, F.; Fernández, M.L.; Maneiro, J.; Pazos, Y.; Salgado, C.; Blanco, J. Accumulation and transformation of DSP toxins in mussels Mytilus galloprovincialis during a toxic episode caused by Dinophysis acuminata. Aquat. Toxicol. 2003, 62, 269–280. [Google Scholar] [CrossRef]
- Gersen, A.; Pol-Hofstad, I.E.; Poelman, M.; Mulder, P.P.; Van den Top, H.; De Boer, J. Marine toxins: Chemistry, toxicity, occurrence and detection, with special reference to the Dutch situation. Toxins 2010, 2, 878–904. [Google Scholar] [CrossRef] [Green Version]
- Blanco, J. Accumulation of Dinophysis toxins in bivalve molluscs. Toxins 2018, 10, 453. [Google Scholar] [CrossRef] [Green Version]
- Manfrin, C.; Dreos, R.; Battistella, S.; Beran, A.; Gerdol, M.; Varotto, L.; Lanfranchi, G.; Venier, P.; Pallavicini, A. Mediterranean mussel gene expression profile induced by okadaic acid exposure. Environ. Sci. Technol. 2010, 44, 8276–8283. [Google Scholar] [CrossRef] [PubMed]
- Dou, M.; Jiao, Y.-H.; Zheng, J.-W.; Zhang, G.; Li, H.-Y.; Liu, J.-S.; Yang, W.-D. De novo transcriptome analysis of the mussel Perna viridis after exposure to the toxic dinoflagellate Prorocentrum lima. Ecotoxicol. Environ. Saf. 2020, 192, 110265. [Google Scholar] [CrossRef]
- Ritz, V.; Marwitz, J.; Sieder, S.; Ziemann, C.; Hirsch-Ernst, K.I.; Quentin, I.; Steinfelder, H.J. Contribution of mdr1b-type P-glycoprotein to okadaic acid resistance in rat pituitary GH3 cells. Naunyn Schmiedebergs Arch. Pharmacol. 1999, 360, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Svenson, S.; Särngren, A.; Förlin, L. Mussel blood cells, resistant to the cytotoxic effects of okadaic acid, do not express cell membrane p-glycoprotein activity (multixenobiotic resistance). Aquat. Toxicol. 2003, 65, 27–37. [Google Scholar] [CrossRef]
- Vale, C.; Botana, L.M. Marine toxins and the cytoskeleton: Okadaic acid and Dinophysis toxins. FEBS J. 2008, 275, 6060–6066. [Google Scholar] [CrossRef] [PubMed]
- Bard, S.M. Multixenobiotic resistance as a cellular defence mechanism in aquatic organisms. Aquat. Toxicol. 2000, 48, 357–389. [Google Scholar] [CrossRef]
- Kurulec, B. The multixenobiotic resistance mechanism in aquatic organisms. Crit. Rev. Toxicol. 1992, 22, 23–43. [Google Scholar] [CrossRef] [PubMed]
- Kurulec, B. A new type of hazardous chemical: The chemosensitizers of multixenobiotic resistance. Environ. Health Perspect. 1997, 105, 855–886. [Google Scholar] [CrossRef] [Green Version]
- Luedeking, A.; Koehler, A. Regulation of expression of multixenobiotic resistance (MXR) genes by environmental factors in the blue mussel Mytilus edulis. Aquat. Toxicol. 2004, 69, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sandrine, P.; Marc, P. Identification of multixenobiotic defence mechanism (MXR) background activities in the freshwater bivalve Dreissena polymorpha as reference values for its use as biomarker in contaminated ecosystems. Chemosphere 2007, 67, 1258–1263. [Google Scholar]
- Martins, J.C.; Domínguez-Pérez, D.; Azevedo, C.; Braga, A.C.; Costa, P.R.; Osório, H.; Vasconcelos, V.; Campos, A. Molecular responses of mussel Mytilus galloprovincialis associated to accumulation and depuration of marine biotoxins okadaic acid and dinophysistoxin-1 revealed by shotgun proteomics. Front. Mar. Sci. 2020, 7, 589822. [Google Scholar] [CrossRef]
- Leslie, E.M.; Deeley, R.G.; Cole, S.P. Multidrug resistance proteins: Role of P-glycoprotein, MRP1, MRP2 and BCRP (ABCG2) in tissue defence. Toxicol. Appl. Pharmacol. 2005, 204, 216–237. [Google Scholar] [CrossRef] [PubMed]
- Szakás, G.; Varadi, A.; Ozvegy-Laczka, C.; Sarkadi, B. The role of ABC transporters in drug absorption, metabolism excretion and toxicity (ADME-Tox). Drug Discov. Today 2008, 13, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Lozano, V.; Martínez-Escauriaza, R.; Pérez-Parallé, M.L.; Pazos, A.J.; Sánchez, J.L. Two novel multidrug resistance associated proteins (MRP/ABCC) from the Mediterranean mussel (Mytilus galloprovincialis): Characterization and expression patterns in detoxifying tissues. Can. J. Zool. 2015, 93, 567–578. [Google Scholar] [CrossRef]
- Ambudkar, S.V.; Dey, S.; Hrycyna, C.A.; Ramachandra, M.; Pastan, I.; Gottesman, M.M. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu. Rev. Pharmacol. Toxicol. 1999, 39, 361–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eufemia, N.A.; Epel, D. Induction of the multixenobiotic defense mechanism (MXR), P-glycoprotein, in the mussel Mytilus californianus as a general cellular response to environmental stresses. Aquat. Toxicol. 2000, 49, 89–100. [Google Scholar] [CrossRef]
- Achard, M.; Baudrimont, M.; Boudou, A.; Bourdineaud, J.P. Induction of a multixenobiotic resistance protein (MXR) in the Asiatic clam Corbicula fluminea after heavy metals exposure. Aquat. Toxicol. 2004, 67, 347–357. [Google Scholar] [CrossRef]
- Hennessy, M.; Spiers, J.P. A primer on the mechanics of P-glycoprotein the multidrug transporter. Pharmacol. Res. 2007, 55, 1–15. [Google Scholar] [CrossRef]
- Xu, Y.-Y.; Liang, J.-J.; Yang, V.-D.; Wang, J.; Li, H.-Y.; Liu, J.-S. Cloning and expression analysis of P-glycoprotein gene in Crassostrea ariakensis. Aquaculture 2014, 418, 39–47. [Google Scholar] [CrossRef]
- Huang, L.; Wang, J.; Chen, W.C.; Li, H.Y.; Liu, J.S.; Jiang, T.; Yang, W.D. P-glycoprotein expression in Perna viridis after exposure to Prorocentrum lima, a dinoflagellate producing DSP toxins. Fish. Shellfish Immunol. 2014, 39, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Liu, S.L.; Zheng, J.-W.; Li, H.-Y.; Liu, J.-S.; Yang, W.-D. P-Glycoprotein and its inducible expression in three bivalve species after exposure to Prorocentrum lima. Aquat. Toxicol. 2015, 169, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Sharom, F.J. The P-glycoprotein multidrug transporter. Essays Biochem. 2011, 50, 161–178. [Google Scholar] [CrossRef] [Green Version]
- Ehlers, A.; These, A.; Hessel, S.; Preiss-Weigert, A.; Lampen, A. Active elimination of the marine biotoxin okadaic acid by P-glycoprotein through an in vitro gastrointestinal barrier. Toxicol. Lett. 2014, 225, 311–317. [Google Scholar] [CrossRef]
- Kamermans, P.; Galley, T.; Boudry, P.; Fuentes, J.; McCombie, H.; Batista, F.M.; Blanco, A.; Dominguez, L.; Cornette, F.; Pincot, L.; et al. Blue mussel hatchery technology in Europe. In Advances in Aquaculture Hatchery Technology; Allan, G.B., Ed.; Elsevier, Woodhead Publishing Series in Food Science, Technology and Nutrition; Woodhead Publishing Limited: Cambridge, UK, 2013; pp. 339–373. [Google Scholar]
- Blanco, J.; Correa, J.; Muñiz, S.; Mariño, C.; Martin, H.; Arévalo, A. Evaluation of the impact of methods and level used for monitoring toxins in mussels. Rev. Galega Recur. Mariños 2013, 3, 1–55. Available online: http://www.recmar-galica.org/pdf/BlancoRGRM_Art_3_2013.pdf (accessed on 22 September 2014).
- Pino-Querido, A.; Álvarez-Castro, J.A.; Guerra-Varela, J.; Toro, M.A.; Vera, M.; Gómez-Pardo, B.; Fuentes, J.; Blanco, J.; Martínez, P. Heritability estimation for okadaic acid algal toxin accumulation, mantle colour and growth traits in Mediterranean mussel (Mytilus galloprovincialis). Aquaculture 2015, 440, 32–39. [Google Scholar] [CrossRef]
- Franzellitti, S.; Fabbri, E. Cytoprotective responses in the Mediterranean mussel exposed to Hg2+ and CH3Hg+. Biochem. Biophys. Res. Commun. 2006, 351, 719–725. [Google Scholar] [CrossRef]
- Ling, D.; Salvaterra, P.M. Robust RT-qPCR data normalization: Validation and selection of internal reference genes during post-experimental data analysis. PLoS ONE 2011, 6, e17762. [Google Scholar] [CrossRef] [Green Version]
- Dean, M.; Hamon, Y.; Chimi, G. The human ATP-binding cassette (ABC) transporter superfamily. J. Lipid. Res. 2001, 42, 1007–1017. [Google Scholar] [CrossRef]
- Ward, A.; Reyes, C.L.; Yu, J.; Roth, C.B. Flexibility in the ABC MsbA alternating access with a twist. Proc. Natl. Acad. Sci. USA 2007, 104, 19005–19010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, J.; Cai, Y.; Pan, L.; Zhen, L. Molecular cloning and characterization of a MXR-related P-glycoprotein cDNA in scallop Chlamys farreri: Transcriptional response to benzo (a) pyrene, tetrabromobisphenol A and endosulfan. Ecotoxicol. Environ. Saf. 2014, 110, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Choudhuri, S.; Klaasen, C.D. Structure, function, expression, genomic organization and single nucleotide polymorphisms of human ABCB1 (MDR1), ABCC (MRP) and ABCG2 (BCRP) efflux transporters. Int. J. Toxicol. 2006, 25, 231–259. [Google Scholar] [CrossRef]
- Prado-Alvarez, M.; Flórez-Barrós, M.F.; Méndez, J.; Fernández-Tajes, J. Effect of okadaic acid on carpet shell clam (Ruditapes decussatus) haemocytes by in vitro exposure and harmful algal bloom simulation assays. Cell Biol. Toxicol. 2013, 29, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Svenson, S.; Förlin, L. Effects of okadaic acid in the blue mussel Mytilus edulis, and rainbow trout Oncorhynchus mydiss. Mar. Environ. Res. 1998, 49, 449–452. [Google Scholar] [CrossRef]
- Bauder, A.; Cembella, A. Viability of the toxic dinoflagellate Prorocentrum lima following ingestion and gut passage in the by scallop Argopecten irradians. J. Shell. Res. 2000, 19, 321–324. [Google Scholar]
- Flórez-Barros, F.; Prado-Álvarez, M.; Méndez, J.; Fernández-Tajes, J. Evaluation of genotoxicity in gills and hemolymph of clam Ruditapes decussatus fed with the toxic dinoflagellate Prorocentrum lima. J. Toxicol. Environ. Health 2011, 74, 971–979. [Google Scholar] [CrossRef] [PubMed]
- Prego-Faraldo, M.V.; Valdiglesias, V.; Méndez, J.; Eirín-López, J.M. Okadaic acid meet and greet: An insight into detection methods, response strategies and genotoxic effects in marine invertebrates. Mar. Drugs 2013, 11, 2829–2845. [Google Scholar] [CrossRef] [Green Version]
- Kington, S.; Chitramvong, Y.; Janvilislri, T. ATP-binding cassette multidrug transporters in Indian-rock oyster Sacrosstrea forskali and their role in the export of an environmental organic pollutant tributyrin. Aquat. Toxicol. 2007, 85, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Shinkel, A.H.; Smit, J.J.; van Tellingen, O.; Beignen, J.H.; Wagenar, E.; van Deenter, L.; Mol, C.A.; van der Valk, M.A.; Robanus-Maandag, E.C.; Riele, H.P. Disruption of the mouse mdr1 a P-glycoprotein gene leads to a deficiency in the blood-brain barrier and to increase sensitivity to drugs. Cell 1994, 77, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Yasumoto, T.; Oshima, Y.; Yamaguchi, M. Occurrence of a new type of shellfish poisoning in the Tohoku district. Bull. Japan. Soc. Sci Fish. 1978, 44, 1249–1255. [Google Scholar] [CrossRef]
- Murata, M.; Shimitami, M.; Sugitani, H.; Oshima, Y.; Yasumoto, T. Isolation and structural elucidation of the causative toxin of the diarrhetic shellfish poisoning. Nippon Susisan Gakkaishi 1982, 48, 549–552. [Google Scholar] [CrossRef] [Green Version]
- Svenson, S. Depuration of okadaic acid (diarreic shellfish toxin) in mussels, Mytilus edulis (Linnaeus), feeding on different quantities of nontoxic algae. Aquaculture 2003, 218, 277–291. [Google Scholar] [CrossRef]
- Suzuki, T.; Igarashi, T.; Ichimi, K.; Watai, M.; Suzuki, M.; Ogiso, E.; Yasumoto, T. Kinetics of diarreic shellfish poisoning toxins, okadaic acid, dinophysistoxin-1, pectenotoxin-6 and yessotoxin in scallops Patinopecten yessoensis. Fisher. Sci. 2005, 71, 948–955. [Google Scholar] [CrossRef]
- Blanco, J.; Mariño, C.; Martin, H.; Acosta, C.P. Anatomical distribution of diarrhetic shellfish poisoning (DSP) toxins in the mussel Mytilus galloprovincialis (Lamarck). Toxicon 2007, 50, 1011–1018. [Google Scholar] [CrossRef]
- McCarron, P.; Kilcoyne, J.; Hess, P. Effects of cooking end heat treatment on concentration and tissue distribution of okadaic acid and Dinophysis toxin-2 in mussels (Mytilus edulis). Toxicon 2008, 51, 1081–1089. [Google Scholar] [CrossRef]
- Paetzold, S.C.; Ross, N.W.; Richards, R.C.; Jones, M.; Hellou, J.; Bard, S.M. Up-regulation of hepatic ABCC2, ABCG2, CYP1A1 and GST in multixenobiotic resistant killifish (Fundulus heteroclitus) from the Sydney Tar Ponds, Nova Scotia Canada. Can. Mar. Environ. Res. 2009, 68, 37–47. [Google Scholar] [CrossRef]
- Epel, D.; Luckenbach, T.; Svenson, C.N.; Macmanus-Spencer, L.A.; Hamdoun, A.; Smital, T. Efflux transporter newly appreciated roles in protection against pollutants. Environ. Sci. Technol. 2008, 6, 3914–3920. [Google Scholar] [CrossRef] [Green Version]
- Van Tellingen, O.; Buckle, J.; Jonker, J.W.; Van der Valk, M.A.; Beijnen, J.H. P-glycoprotein and Mrp1 collectively protect the bone marrow from vincristine-induced toxicity in vivo. Br. J. Cancer 2003, 89, 1776–1782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.P.; Johnson, D.R.; Finch, R.A.; Belinsky, M.G.; Kruh, G.D.; Sartorelli, A.C. Comparative study of the importance of multidrug resistance-associated protein 1 and P-glycoprotein to drug sensitivity in immortalised mouse embryonic fibroblasts. Mol. Cancer Ther. 2002, 1, 1105–1114. [Google Scholar]
- Ye, Z.; Lu, Y.; Wu, T. The impact of ATP-binding cassette transporters on metabolic diseases. Nutr. Metab. 2020, 17, 61. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Wootton, J.C.; Gertz, E.M.; Agarwala, R.; Morgulis, A.; Schäffer, A.A.; Yu, Y.-K. Protein database searches using compositionally adjusted substitutions matrices. FEBS J. 2005, 272, 5101–5109. [Google Scholar] [CrossRef]
- Pearson, W.R.; Lipman, D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 1988, 85, 2444–2448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; López, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [Green Version]
- Mauriz, O.; Maneiro, V.; Pérez-Parallé, M.L.; Sánchez, J.L.; Pazos, A.J. Selection of reference genes for quantitative RT-PCR studies on the gonad of the bivalve mollusc Pecten maximus L. Aquaculture 2012, 370, 158–165. [Google Scholar] [CrossRef]
- Vale, P.; Sampayo, M.A. Esters of okadaic acid and dinophysistoxin-2 in Portuguese bivalves related to human poisonings. Toxicon 1999, 37, 1109–1121. [Google Scholar] [CrossRef]
- Regueiro, J.; Rossignoli, A.E.; Álvarez, G.; Blanco, J. Automated on-line solid phase extraction coupled to liquid chromatography–tandem mass spectrometry for determination of lipophilic marine toxins in shellfish. Food Chem. 2011, 129, 533–540. [Google Scholar] [CrossRef] [PubMed]



| Primer Name | Sequence (5′-3′) | Target | Amplicon (bp) | Annealing T |
|---|---|---|---|---|
| mdr1 | ||||
| MDR1F | CAGAGGTTCTATGACCCAGATGCAG | RT-PCR amplification | 381 | 54.5 °C |
| MDR1R | GTTCTCACTCTCAGAGTCTAATGCAG | |||
| MDR1F2 | CCATTGCCAGAGCTTTGATYAGAGACCC | 3′Race PCR | 1067 | 65 °C |
| MDR1R2 | CTGYCCTCCYGATAACTGAGCTCCAYGC | 5′Race PCR | 1830 | 65 °C |
| MDRMgFw | GTAGCAGCCCTGGTTATAGC | 3′Race PCR | 1463 | 64.9 °C |
| MDR End Nested3′ | GTAGAAAGTGGTACACACCAGACTC | 3′Race PCR | 217 | 65 °C |
| mdr2 | ||||
| MDR2 Start Rc3′F | TGCCAGAACCAGAATTATTGGGAT | RT-PCR amplification (+qRt Rv) | 1290 | 50–56 °C |
| MDR2 Race3′ Fw 1 | AAGAAGGGAGAAGAGGAAGAAAAGGA | 3′Race PCR/RT-PCR amplif. (+Rv1&Rv2) | 850/1161–1249 | 65 °C/55 °C |
| MDR2 Race3′ Fw 2 | TCGTTATAACGCTCCTGAATGGCC | 3′Race PCR | 475 | 65 °C |
| MDR2 Race3′ Fw 3 | TGGTTGGATGTATTGCTGCATGTTTGA | RT-PCR amplification + (Rv1&Rv2) | 1070–1158 | 55 °C |
| MDR2 Race3′ Fw 4 | GTGTTTGGAGCTATGGCTTTAGGACA | RT-PCR amplification (+Ultimo2 Rv) | 1117 | 58 °C |
| MDR2 qRT Fw | GAGCCAAACTGGTAAGAGAGG | 3′Race PCR | 1082 | 65 °C |
| MDR2 qRT Rv | GTGGTGGAGCAACATTACCA | RT-PCR amplification (+Start Rc3′ F) | 50–56 °C | |
| MDR2 Fr Fw | TGGTGAGAGAGGAGCCCAGC | RT-PCR amplification | 640 | 58 °C |
| MDR2 Fr Rv | AAATGCTGGCTGAACTCCAC | |||
| MDR2 Rv1 | TGCCAGTGTCTGTCCTGGATC | RT-PCR amplification (+Race3′ Fw 1& Fw 4) | 55 °C | |
| MDR2 Rv2 | GGCAGGATCTGTGTTGCTTG | RT-PCR amplification (+Race3′ Fw 1& Fw 4) | 55 °C | |
| MDR2 End2 Rv | GCACTTACAATACACAGCAC | RT-PCR amplification (+Race Fw 4) | 65 °C | |
| MDR2Fin R3′F | AGCTGCTAGGAACGCTAACATTCATG | 3′Race PCR | 591 | 65 °C |
| Gene | Sense Primer (5′–3′) | Antisense Primer (5′–3′) | Amplicon Length | E% (DG) | E% (GI) | E% (MT) |
|---|---|---|---|---|---|---|
| mdr1 | GTGGGCTCTAGCTCTTGTTG | GTCTTCCCAGCCTCCTCTAG | 126 | 100.1 | 111.6 | 108.9 |
| mdr2 | TGGAGCCTATGCTCTTGGG | CAACATTACCAATGGACCACGC | 131 | 99.8 | 105.1 | 91.7 |
| nd4 | CAGCCCCACCTAGTCTAAATC | AGCAAGCCCTAATAAAGCTCATC | 114 | 105.8 | 100.5 | 100.9 |
| gapdh | AGGAATGGCCTTCAGGGTAC | TCAGATGCTGCTTTAATGGCTG | 114 | 99.3 | 107.3 | 96.5 |
| cox1 | TGCTCATTGGCATTGGGTGTC | AGTTCCTGCTCAGTCCATCTCAC | 151 | 85.9 | 97.6 | 91.6 |
| rps27 | CGTGAATGTCCCAACGAAGAG | TGTTGCCTCTGGTTTGTTGA | 114 | 92 | 101.2 | 97.3 |
| tif5a | ACGCTACTTGACATTAACGATG | AGCTAGTTCTTCTCCCATAGC | 171 | 96.9 | 99.3 | 104.6 |
| rps4 | TGGGTTATCGAGGGCGTAG | TCCCTTAGTTTGTGAGGACCTG | 121 | 91.1 | 93.5 | 95.8 |
| act | TCTTGATTTCGAGCAGGAAATG | GGATGGTTGGAATAATGATTCTG | 138 | 91 | 111.3 | 100.3 |
| 18S | TCGATGGTACGTGATATGCC | CGTTTCTCATGCTCCCTCTC | 84 | 99.2 | 87.7 | 94 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Escauriaza, R.; Lozano, V.; Pérez-Parallé, M.L.; Blanco, J.; Sánchez, J.L.; Pazos, A.J. Expression Analyses of Genes Related to Multixenobiotic Resistance in Mytilus galloprovincialis after Exposure to Okadaic Acid-Producing Dinophysis acuminata. Toxins 2021, 13, 614. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13090614
Martínez-Escauriaza R, Lozano V, Pérez-Parallé ML, Blanco J, Sánchez JL, Pazos AJ. Expression Analyses of Genes Related to Multixenobiotic Resistance in Mytilus galloprovincialis after Exposure to Okadaic Acid-Producing Dinophysis acuminata. Toxins. 2021; 13(9):614. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13090614
Chicago/Turabian StyleMartínez-Escauriaza, Roi, Vanessa Lozano, M. Luz Pérez-Parallé, Juan Blanco, José L. Sánchez, and Antonio J. Pazos. 2021. "Expression Analyses of Genes Related to Multixenobiotic Resistance in Mytilus galloprovincialis after Exposure to Okadaic Acid-Producing Dinophysis acuminata" Toxins 13, no. 9: 614. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13090614