Identification of Fish Species and Toxins Implicated in a Snapper Food Poisoning Event in Sabah, Malaysia, 2017
Abstract
:1. Introduction
2. Results
2.1. Incident Case
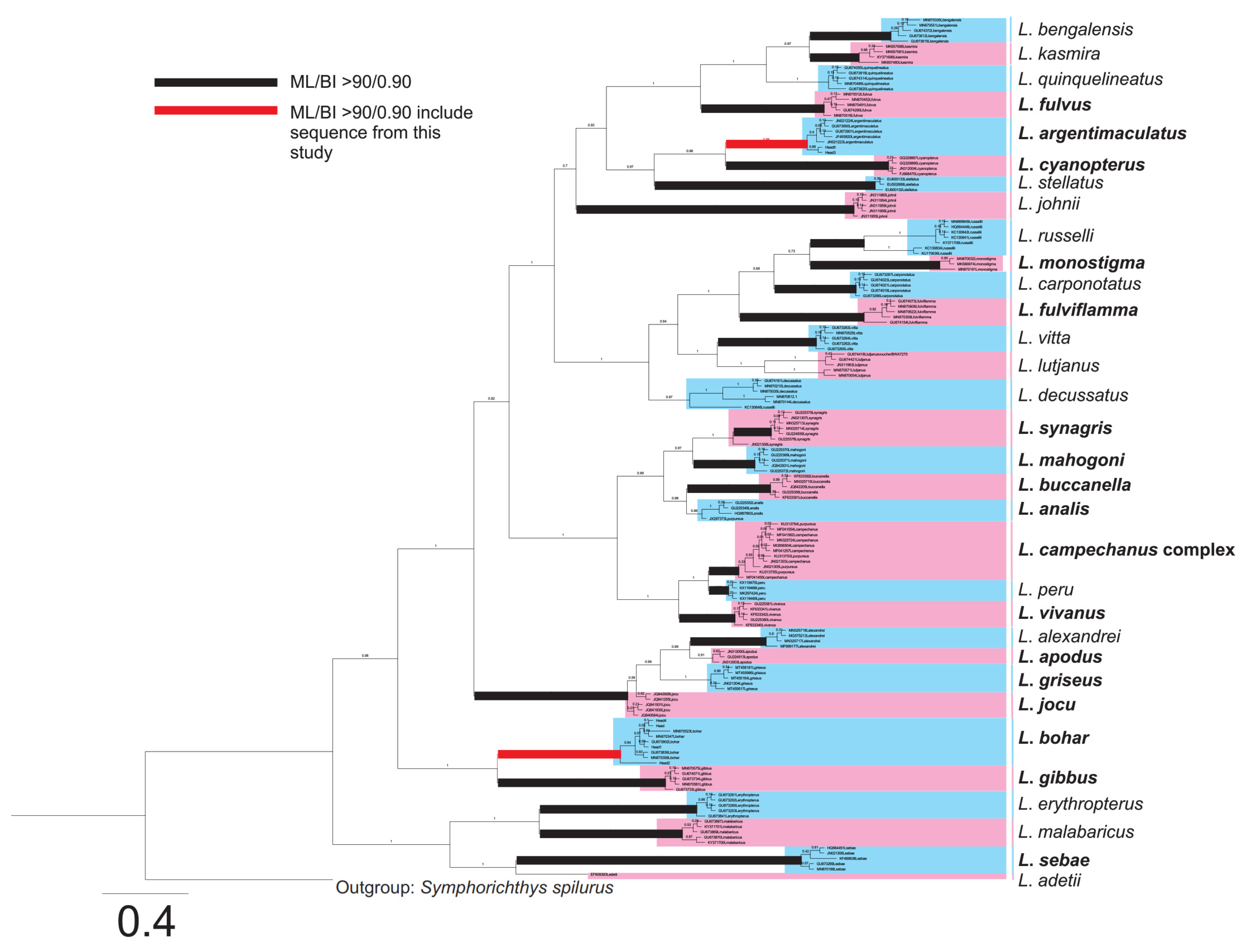
2.2. Species Identification of Fish Samples
2.3. Toxin in Fish Samples
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Reagents and Standards
4.3. Molecular Characterization of Fish Samples
4.3.1. DNA Extraction and PCR
4.3.2. Taxon Sampling and Sequence Alignment
4.3.3. Pairwise Genetic Distance and Phylogenetic Analysis
4.4. Extraction and Clean-Up of Fish Samples for LC/MS
4.5. Quadrupole LC/MS
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Silva, M.; Rodriguez, I.; Barreiro, A.; Kaufmann, M.; Neto, A.I.; Hassouani, M.; Sabour, B.; Alfonso, A.; Botana, L.M.; Vasconcelos, V. First report of ciguatoxins in two starfish species: Ophidiaster ophidianus and Marthasterias glacialis. Toxins 2015, 7, 3740–3757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roué, M.; Darius, H.T.; Picot, S.; Ung, A.; Viallon, J.; Gaertner-Mazouni, N.; Sibat, M.; Amzil, Z.; Chinain, M. Evidence of the bioaccumulation of ciguatoxins in giant clams (Tridacna maxima) exposed to Gambierdiscus spp. cells. Harmful Algae 2016, 57, 78–87. [Google Scholar] [CrossRef] [Green Version]
- Darius, H.T.; Roué, M.; Sibat, M.; Viallon, J.; Gatti, C.M.i.; Vandersea, M.W.; Tester, P.A.; Litaker, R.W.; Amzil, Z.; Hess, P.; et al. Tectus niloticus (Tegulidae, Gastropod) as a novel vector of ciguatera poisoning: Detection of Pacific ciguatoxins in toxic samples from Nuku Hiva Island (French Polynesia). Toxins 2018, 10, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, T.Y.K. Ciguatera fish poisoning in East Asia and Southeast Asia. Mar. Drugs 2015, 13, 3466–3478. [Google Scholar] [CrossRef] [Green Version]
- Toda, M.; Uneyama, C.; Toyofuku, H.; Morikawa, K. Trends of food poisonings caused by natural toxins in Japan, 1989–2011. Shokuhin eiseigaku zasshi. J. Food Hyg. Soc. Jpn. 2012, 53, 105–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrero, E.T. Fish Poisoning in Basilan. Available online: http://seafoodtoxinmedic.tripod.com/HTMLobj-119/FISHPOISONINGINBASILAN.doc (accessed on 26 February 2015).
- Chen, T.Y.; Chen, N.H.; Lin, W.F.; Hwang, K.L.; Huang, Y.C.; Hwang, D.F. Identification of causative fish for a food poisoning in Taiwan by using SDS-PAGE technique. J. Mar. Sci. Technol. 2010, 18, 593–596. [Google Scholar] [CrossRef]
- Tester, P.A.; Kibler, S.R.; Holland, W.C.; Usup, G.; Vandersea, M.W.; Leaw, C.P.; Teen, L.P.; Larsen, J.; Mohammad-Noor, N.; Faust, M.A.; et al. Sampling harmful benthic dinoflagellates: Comparison of artificial and natural substrate methods. Harmful Algae 2014, 39, 8–25. [Google Scholar] [CrossRef]
- Dai, X.; Mak, Y.L.; Lu, C.-K.; Mei, H.-H.; Wu, J.J.; Lee, W.H.; Chan, L.L.; Lim, P.T.; Mustapa, N.I.; Lim, H.C. Taxonomic assignment of the benthic toxigenic dinoflagellate Gambierdiscus sp. type 6 as Gambierdiscus balechii (Dinophyceae), including its distribution and ciguatoxicity. Harmful Algae 2017, 67, 107–118. [Google Scholar] [CrossRef]
- Yong, H.L.; Mustapa, N.I.; Lee, L.K.; Lim, Z.F.; Tan, T.H.; Usup, G.; Gu, H.; Litaker, R.W.; Tester, P.A.; Lim, P.T. Habitat complexity affects benthic harmful dinoflagellate assemblages in the fringing reef of Rawa Island, Malaysia. Harmful Algae 2018, 78, 56–68. [Google Scholar] [CrossRef]
- Usup, G. Seafood toxicity in Malaysia (of seafood, mice and man). In Proceedings of the 7th Kelantan Helath Conference, Kota Bharu, Malaysia, 15–16 June 2011. [Google Scholar]
- Nik Khairol Reza, B.M.Y.; Wan Mansor, B.H.; Anita, B.S.; Fauziah, B.M.N.; Mat Ghani, B.M.; Sahari, B.C.H.; Noor Iznina, B.A.A. Ciguatera poisoning after imported red sanpper fish ingestion in Jeli, Kelantan, Malaysia, 8–10 September 2010. In Proceedings of the 7th Kelantan Helath Conference, Kota Bharu, Malaysia, 15–16 June 2011. [Google Scholar]
- Lee, H.G.; Leaw, C.P.; Lim, P.; Jipanin, S. Ciguatera fish poisoning: First reported case in Sabah, Malaysia. Med. J. Malays. 2019, 74, 545–546. [Google Scholar]
- Kawakami, T.; Aoyama, J.; Tsukamoto, K. Morphology of pelagic fish eggs identified using mitochondrial DNA and their distribution in waters west of the Mariana Islands. Environ. Biol. Fishes 2010, 87, 221–235. [Google Scholar] [CrossRef]
- Yaqub, A.; Kamran, M.; Malkani, N.; Anjum, K.; Faheem, M.; Iqbal, M.; Khan, R. Mitochondrial COI gene based molecular identification and phylogenetic analysis in exotic fish (Oreochromis mossambicus) of Pakistan. J. Anim. Plant Sci. 2019, 29, 1501–1518. [Google Scholar]
- Wibowo, A.; Wahlberg, N.; Vasemägi, A. DNA barcoding of fish larvae reveals uncharacterised biodiversity in tropical peat swamps of New Guinea, Indonesia. Mar. Freshw. Res. 2017, 68, 1079–1087. [Google Scholar] [CrossRef]
- Anjali, K.; Mandal, A.; Gunalan, B.; Ruban, L.; Anandajothi, E.; Thineshsanthar, D.; Manojkumar, T.; Kandan, S. Identification of six grouper species under the genus Epinephelus (Bloch, 1793) from Indian waters using PCR-RFLP of cytochrome c oxidase I (COI) gene fragment. Food Control 2019, 101, 39–44. [Google Scholar] [CrossRef]
- Froese, R.; Pauly, D. FishBase. World Wide Web Electronic Publication. Available online: www.fishbase.org (accessed on 8 July 2021).
- Banner, A.H.; Helfrich, P.; Piyakarnchana, T. Retention of ciguatera toxin by the red snapper, Lutjanus bohar. Copeia 1966, 1966, 297–301. [Google Scholar] [CrossRef]
- Bourdeau, P.; Bagnis, R. Risk factors of ciguatera in the French West Indies in Saint-Barthélémy, Saint-Martin and Anguilla. Rev. Elev. Med. Vet. Pays Trop. 1989, 42, 393–410. [Google Scholar] [PubMed]
- Clausing, R.; Chinain, M.; Dechraoui Bottein, M.-Y. Practical sampling guidance for determination of ciguatoxin in fish. In Guide for Designing and Implementing a Plan to Monitor Toxin-Producing Microalgae; Reguera, B., Alonso, R., Moreira, Á., Méndez, S., Dechraoui Bottein, M.-Y., Eds.; Unesco & IAEA: Paris, France; Vienna, Austria, 2016; pp. 51–63. [Google Scholar]
- Davin, W.T. The Effects of Ciguatoxic Food on Selected Fishes (Ciguatera, Gambierdiscus Toxicus). Ph.D. Dissertation, Southern Illinois University, Carbondale, IL, USA, 1986. [Google Scholar]
- De Motta, G.E.; Feliu, J.; Izquierdo, A. Identification and epidemiological analysis of ciguatera cases in Puerto Rico. Mar. Fish. Rev. 1986, 48, 14–18. [Google Scholar]
- de Sylva, D.P. Distribution and ecology of ciguatera fish poisoning in Florida, with emphasis on the Florida Keys. Bull. Mar. Sci. 1994, 54, 944–954. [Google Scholar]
- Gillespie, N.C.; Lewis, R.J.; Pearn, J.H.; Bourke, A.T.; Holmes, M.J.; Bourke, J.B.; Shields, W.J. Ciguatera in Australia. Occurrence, clinical features, pathophysiology and management. Med. J. Aust. 1986, 145, 584–590. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Konosu, S.; Yasumoto, T.; Kamiya, H. Ciguatera in the Ryukyu and Amami Islands. Bull. Jpn. Soc. Sci. Fish. 1969, 35, 316–326. [Google Scholar] [CrossRef] [Green Version]
- Nordström, S. Ciguatera Fish Poisoning. Master’s Thesis, Uppsala University, Uppsala, Sweden, 2013. [Google Scholar]
- Olsen, D.A.; Nellis, D.W.; Wood, R.S. Ciguatera in the eastern Caribbean. Mar. Fish. Rev. 1984, 46, 13–18. [Google Scholar]
- Randall, J.E. A Review of ciguatera, tropical fish poisoning, with a tentative explanation of its Cause. Bull. Mar. Sci. 1958, 8, 236–267. [Google Scholar]
- Wong, C.-K.; Hung, P.; Lee, K.L.H.; Kam, K.-M. Study of an outbreak of ciguatera fish poisoning in Hong Kong. Toxicon 2005, 46, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.V.; Uesugi, A.; Uchida, H.; Ky, P.X.; Minh, D.Q.; Watanabe, R.; Matsushima, R.; Oikawa, H.; Nagai, S.; Iwataki, M.; et al. Identification of causative ciguatoxins in red snappers Lutjanus bohar implicated in ciguatera fish poisonings in Vietnam. Toxins 2018, 10, 420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yogi, K.; Oshiro, N.; Inafuku, Y.; Hirama, M.; Yasumoto, T. Detailed LC-MS/MS analysis of ciguatoxins revealing distinct regional and species characteristics in fish and causative alga from the Pacific. Anal. Chem. 2011, 83, 8886–8891. [Google Scholar] [CrossRef]
- Suzuki, T.; Ha, D.V.; Uesugi, A.; Uchida, H. Analytical challenges to ciguatoxins. Curr. Opin. Food Sci. 2017, 18, 37–42. [Google Scholar] [CrossRef]
- FAO. Sixth World Congress on Seafood Safety, Quality and Trade. Sydney, Australia, 14–16 September 2005; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2007; p. 217. [Google Scholar]
- Lewis, R.J. The changing face of ciguatera. Toxicon 2001, 39, 97–106. [Google Scholar] [CrossRef]
- Vilariño, N.; Louzao, M.C.; Abal, P.; Cagide, E.; Carrera, C.; Vieytes, M.R.; Botana, L.M. Human poisoning from marine toxins: Unknowns for optimal consumer protection. Toxins 2018, 10, 324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutherland, S.K. Australian Animal Toxins; Oxford University Press: Melbourne, Australia, 1993; p. 527. [Google Scholar]
- Laurent, D.; Yeeting, B.; Labrosse, P.; Gaudechoux, J.-P. Ciguatera: A Field Reference Guide; Agdex Pacific Islands 493/096; Secretariat of the Pacific Community (SPC) and Institute of Research for Development (IRD): Noumea, New Caledonia, 2005; p. 91. Available online: https://coastfish.spc.int/component/content/article/340-ciguatera-field-reference-guide.html (accessed on 1 August 2021).
- Stewart, I.; Eaglesham, G.K.; Poole, S.; Graham, G.; Paulo, C.; Wickramasinghe, W.; Sadler, R.; Shaw, G.R. Establishing a public health analytical service based on chemical methods for detecting and quantifying Pacific ciguatoxin in fish samples. Toxicon 2010, 56, 804–812. [Google Scholar] [CrossRef]
- Mak, Y.L.; Wu, J.J.; Chan, W.H.; Murphy, M.B.; Lam, J.C.W.; Chan, L.L.; Lam, P.K.S. Simultaneous quantification of Pacific ciguatoxins in fish blood using liquid chromatography–tandem mass spectrometry. Anal. Bioanal. Chem. 2013, 405, 3331–3340. [Google Scholar] [CrossRef]
- Wong, C.-K.; Hung, P.; Lo, J.Y.C. Ciguatera fish poisoning in Hong Kong–A 10-year perspective on the class of ciguatoxins. Toxicon 2014, 86, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Kohli, G.S.; Haslauer, K.; Sarowar, C.; Kretzschmar, A.L.; Boulter, M.; Harwood, D.T.; Laczka, O.; Murray, S.A. Qualitative and quantitative assessment of the presence of ciguatoxin, P-CTX-1B, in spanish mackerel (Scomberomorus commerson) from waters in New South Wales (Australia). Toxicol. Rep. 2017, 4, 328–334. [Google Scholar] [CrossRef]
- Murata, M.; Legrand, A.M.; Ishibashi, Y.; Fukui, M.; Yasumoto, T. Structures and configurations of ciguatoxin from the moray eel Gymnothorax javanicus and its likely precursor from the dinoflagellate Gambierdiscus toxicus. J. Am. Chem. Soc. 1990, 112, 4380–4386. [Google Scholar] [CrossRef]
- Satake, M.; Fukui, M.; Legrand, A.-M.; Cruchet, P.; Yasumoto, T. Isolation and structures of new ciguatoxin analogs, 2,3-dihydroxyCTX3C and 51-hydroxyCTX3C, accumulated in tropical reef fish. Tetrahedron Lett. 1998, 39, 1197–1198. [Google Scholar] [CrossRef]
- Lee, L.K.; Lim, Z.F.; Gu, H.; Chan, L.L.; Litaker, R.W.; Tester, P.A.; Leaw, C.P.; Lim, P.T. Effects of substratum and depth on benthic harmful dinoflagellate assemblages. Sci. Rep. 2020, 10, 11251. [Google Scholar] [CrossRef]
- Leaw, C.P.; Tan, T.H.; Lim, H.C.; Teng, S.T.; Yong, H.L.; Smith, K.F.; Rhodes, L.; Wolf, M.; Holland, W.C.; Vandersea, M.W. New scenario for speciation in the benthic dinoflagellate genus Coolia (Dinophyceae). Harmful Algae 2016, 55, 137–149. [Google Scholar] [CrossRef]
- Veneza, I.; Silva, R.D.; Silva, D.D.; Gomes, G.; Sampaio, I.; Schneider, H. Multiloci analyses suggest synonymy among Rhomboplites, Ocyurus and Lutjanus and reveal the phylogenetic position of Lutjanus alexandrei (Lutjanidae: Perciformes). Neotrop. Ichthyol. 2019, 17, e180109. [Google Scholar] [CrossRef]
- Kim, G.; Lee, J.-H.; Alam, M.J.; Lee, S.R.; Andriyono, S. Complete mitochondrial genome of Spanish flag snapper, Lutjanus carponotatus (Perciformes: Lutjanidae). Mitochondrial DNA Part B 2019, 4, 568–569. [Google Scholar] [CrossRef] [Green Version]
- Andriyono, S.; Alam, J.; Kwak, D.H.; Kim, H.-W. Complete mitochondrial genome of brownstripe red snapper, Lutjanus vitta (Perciformes: Lutjanidae). Mitochondrial DNA Part B 2018, 3, 1129–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afriyie, G.; Wang, Z.; Dong, Z.; Ayisi Larbi, C.; Asiedu, B.; Guo, Y. Complete mitochondrial genome and assembled DNA barcoding analysis of Lutjanus fulgens (Valenciennes, 1830) and its comparison with other Lutjanus species. Ecol. Evol. 2020, 10, 7971–7980. [Google Scholar] [CrossRef]
- Gomes, G.; Schneider, H.; Vallinoto, M.; Santos, S.; Orti, G.; Sampaio, I. Can Lutjanus purpureus (South red snapper) be” legally” considered a red snapper (Lutjanus campechanus)? Genet. Mol. Biol. 2008, 31, 372–376. [Google Scholar] [CrossRef] [Green Version]
- da Silva, R.; Pedraza-Marrón, C.D.R.; Sampaio, I.; Betancur-R, R.; Gomes, G.; Schneider, H. New insights about species delimitation in red snappers (Lutjanus purpureus and L. campechanus) using multilocus data. Mol. Phylogen. Evol. 2020, 147, 106780. [Google Scholar] [CrossRef] [PubMed]
- Hubert, N.; Hanner, R.; Holm, E.; Mandrak, N.E.; Taylor, E.; Burridge, M.; Watkinson, D.; Dumont, P.; Curry, A.; Bentzen, P.; et al. Identifying Canadian freshwater fishes through DNA Barcodes. PLoS ONE 2008, 3, e2490. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Oshiro, N.; Yogi, K.; Asato, S.; Sasaki, T.; Tamanaha, K.; Hirama, M.; Yasumoto, T.; Inafuku, Y. Ciguatera incidence and fish toxicity in Okinawa, Japan. Toxicon 2010, 56, 656–661. [Google Scholar] [CrossRef] [PubMed]

| Sample Name | Condition of Sample | Fish Species |
|---|---|---|
| Head 1 | Uncooked, frozen | Lutjanus bohar |
| Head 2 | Uncooked, frozen | Lutjanus bohar |
| Head 3 | Uncooked, frozen | Lutjanusargentimaculatus |
| Head 4 | Uncooked, frozen | Lutjanus bohar |
| Head 5 | Uncooked, frozen | Lutjanusargentimaculatus |
| Flesh 1 | Cooked, partially eaten | Lutjanus |
| Flesh 2 | Cooked, partially eaten | Lutjanus |
| Flesh 3 | Cooked, partially eaten | Lutjanus |
| Tail 1 | Cooked | Lutjanus |
| Sample Name | Fish Species | CTX-1B (ng/g) | Total Toxicity (MU/100g Fish Tissue) Estimated by LC/MS *1 |
|---|---|---|---|
| Head 1 | Lutjanus bohar | ND *2 | - |
| Head 2 | Lutjanus bohar | 0.38 | 5.40 |
| Head 3 | Lutjanus argentimaculatus | ND | - |
| Head 4 | Lutjanus bohar | ND | - |
| Head 5 | Lutjanus argentimaculatus | Trace *3 | - |
| Flesh 1 | Lutjanus | ND | - |
| Flesh 2 | Lutjanus | ND | - |
| Flesh 3 | Lutjanus | ND | - |
| Tail 1 | Lutjanus | ND | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dao, H.V.; Uesugi, A.; Uchida, H.; Watanabe, R.; Matsushima, R.; Lim, Z.F.; Jipanin, S.J.; Pham, K.X.; Phan, M.-T.; Leaw, C.P.; et al. Identification of Fish Species and Toxins Implicated in a Snapper Food Poisoning Event in Sabah, Malaysia, 2017. Toxins 2021, 13, 657. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13090657
Dao HV, Uesugi A, Uchida H, Watanabe R, Matsushima R, Lim ZF, Jipanin SJ, Pham KX, Phan M-T, Leaw CP, et al. Identification of Fish Species and Toxins Implicated in a Snapper Food Poisoning Event in Sabah, Malaysia, 2017. Toxins. 2021; 13(9):657. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13090657
Chicago/Turabian StyleDao, Ha Viet, Aya Uesugi, Hajime Uchida, Ryuichi Watanabe, Ryoji Matsushima, Zhen Fei Lim, Steffiana J. Jipanin, Ky Xuan Pham, Minh-Thu Phan, Chui Pin Leaw, and et al. 2021. "Identification of Fish Species and Toxins Implicated in a Snapper Food Poisoning Event in Sabah, Malaysia, 2017" Toxins 13, no. 9: 657. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13090657





