A Novel Glutathione S-Transferase Gtt2 Class (VpGSTT2) Is Found in the Genome of the AHPND/EMS Vibrio parahaemolyticus Shrimp Pathogen
Abstract
:1. Introduction
2. Results
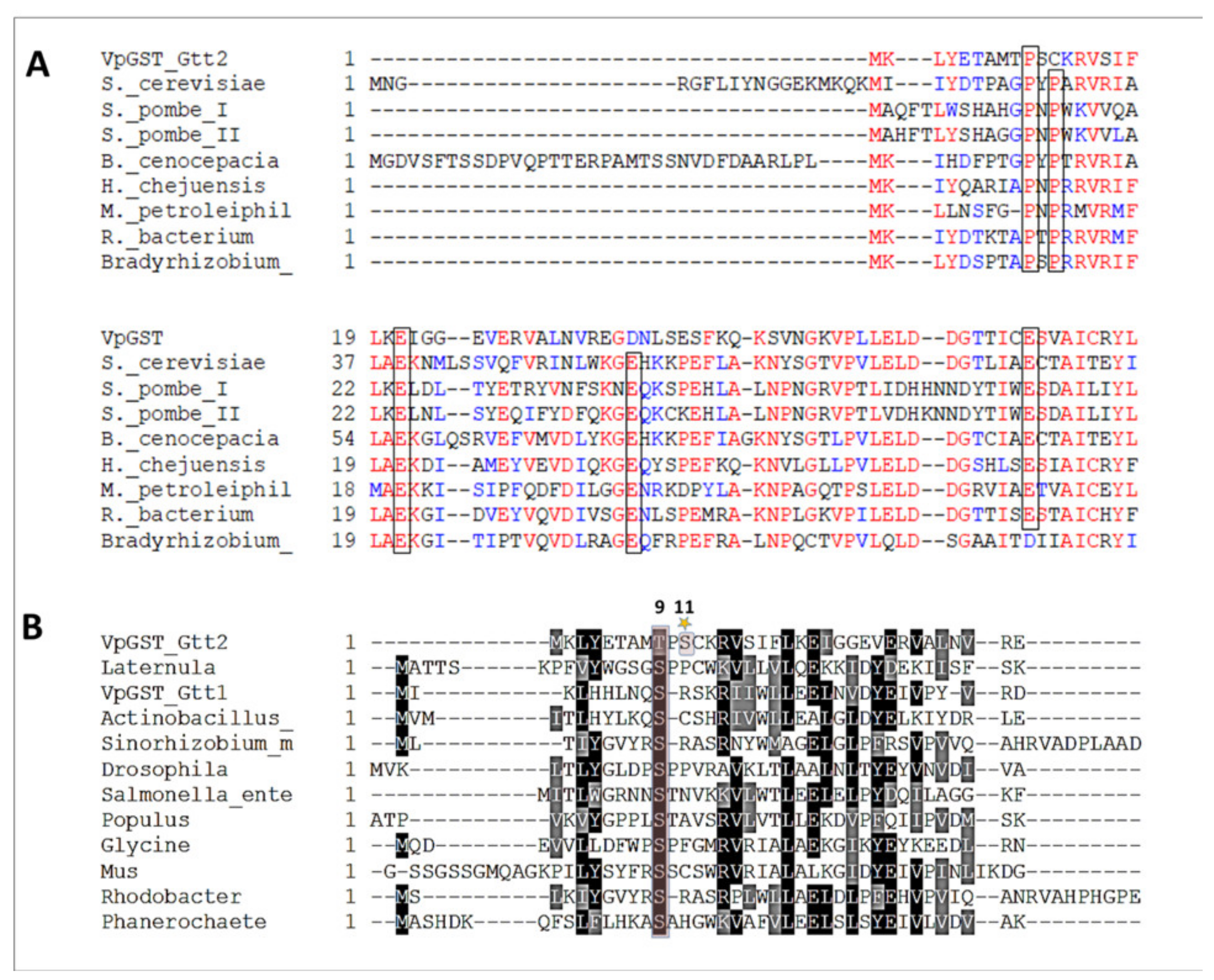
2.1. Phylogenetic and Sequence Analysis of GSTs
2.2. Overexpression and Purification of VpGSTT2
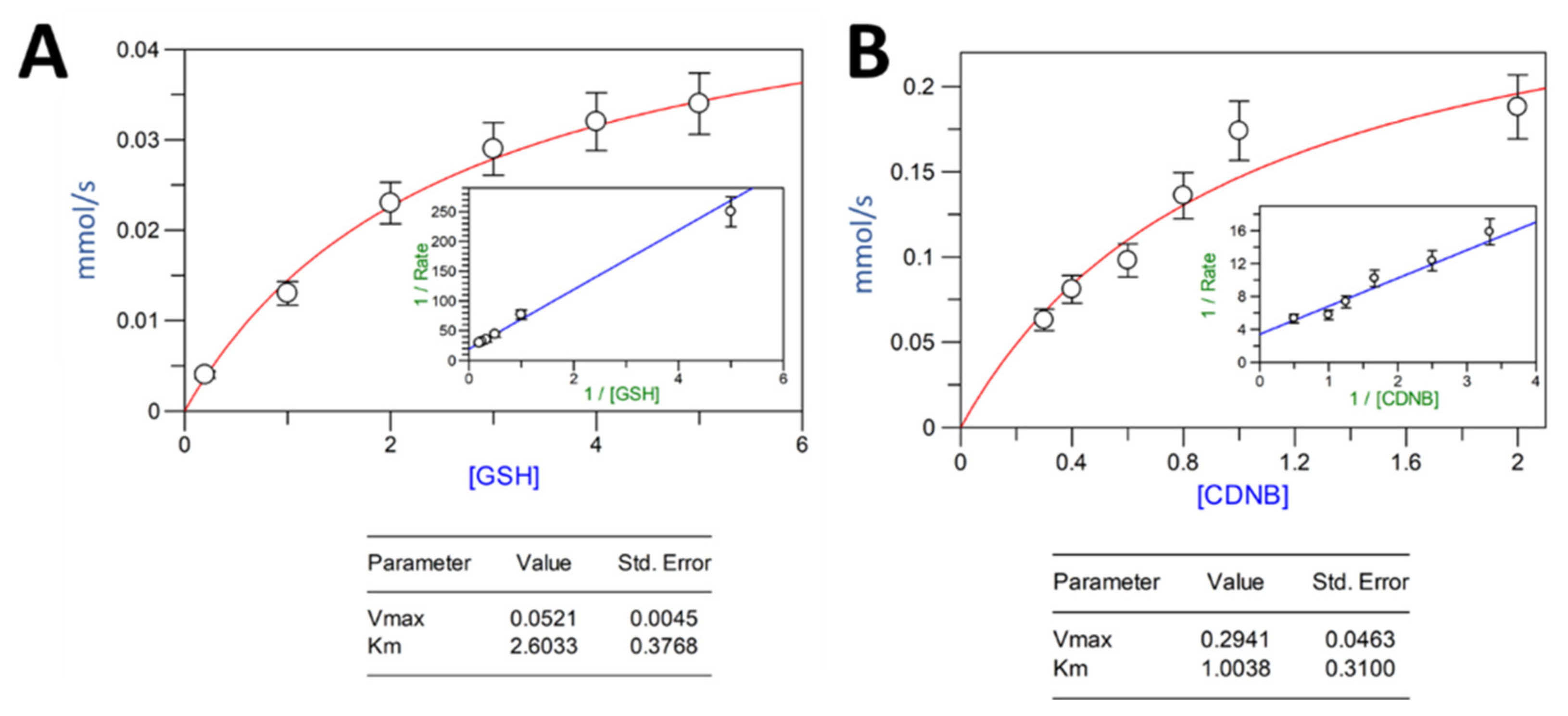
2.3. Activity Assay and Parameter Kinetics for VpGSTT2
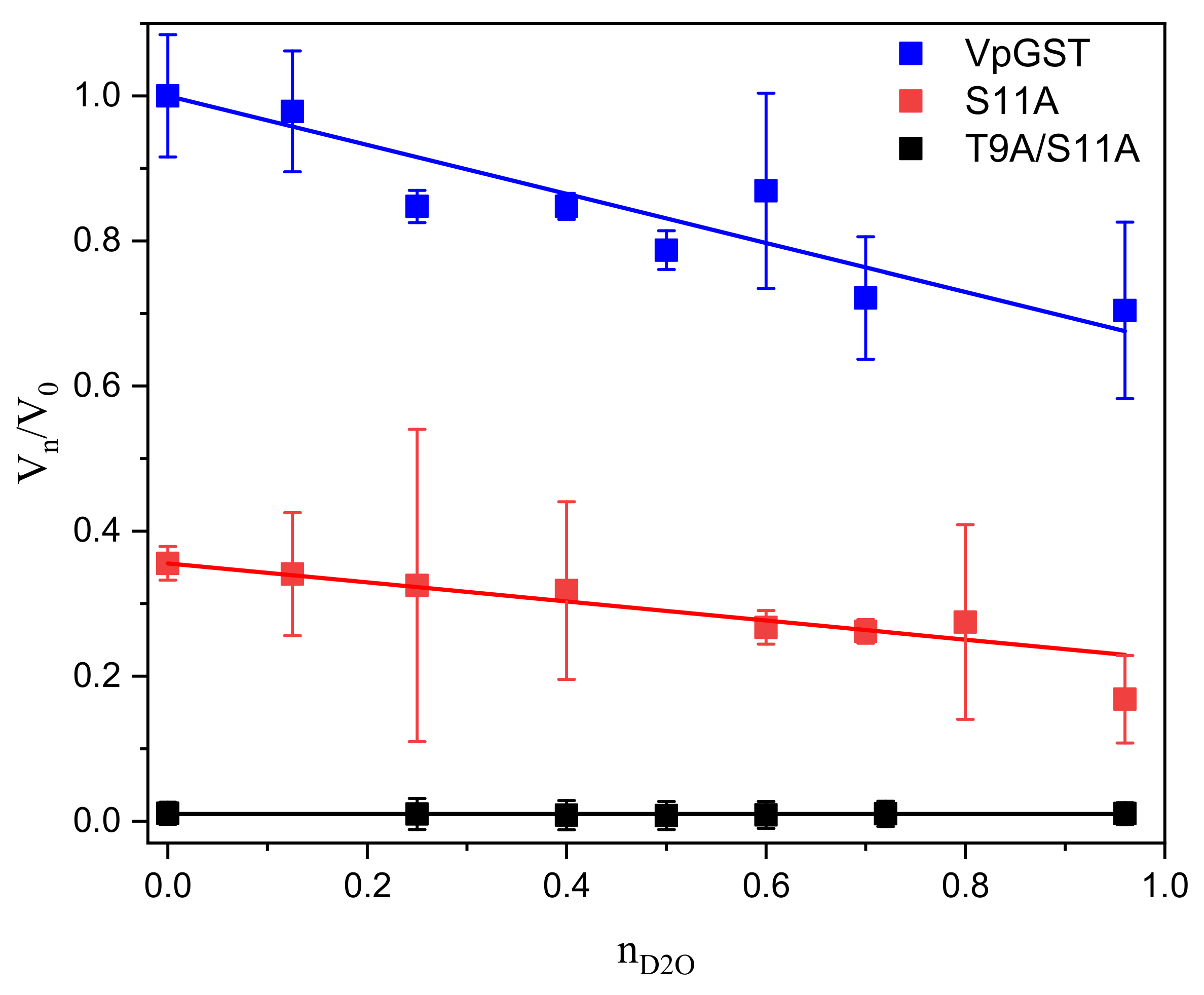
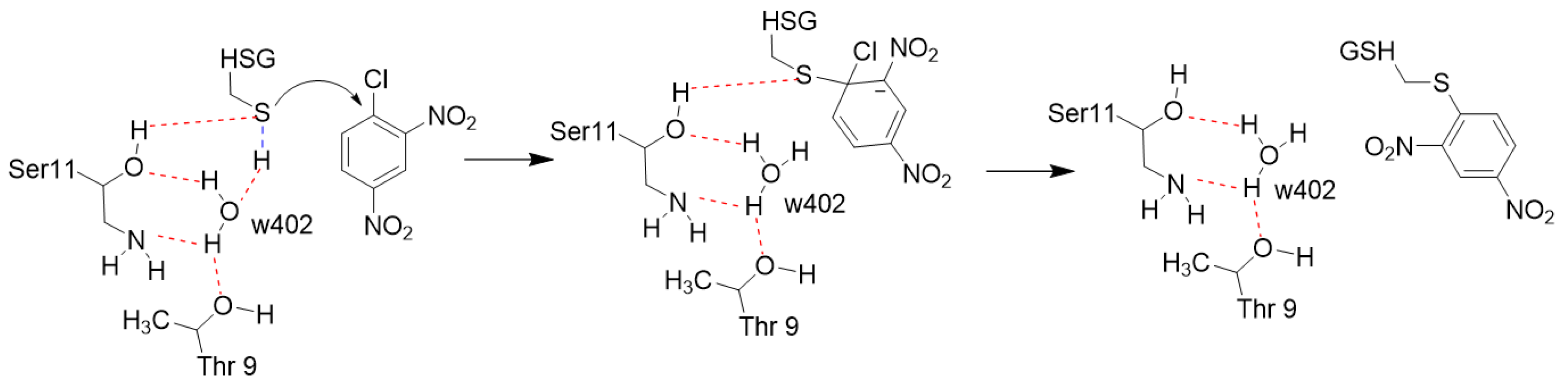
2.4. Solvent Kinetic Isotope Effects: Proton Inventory
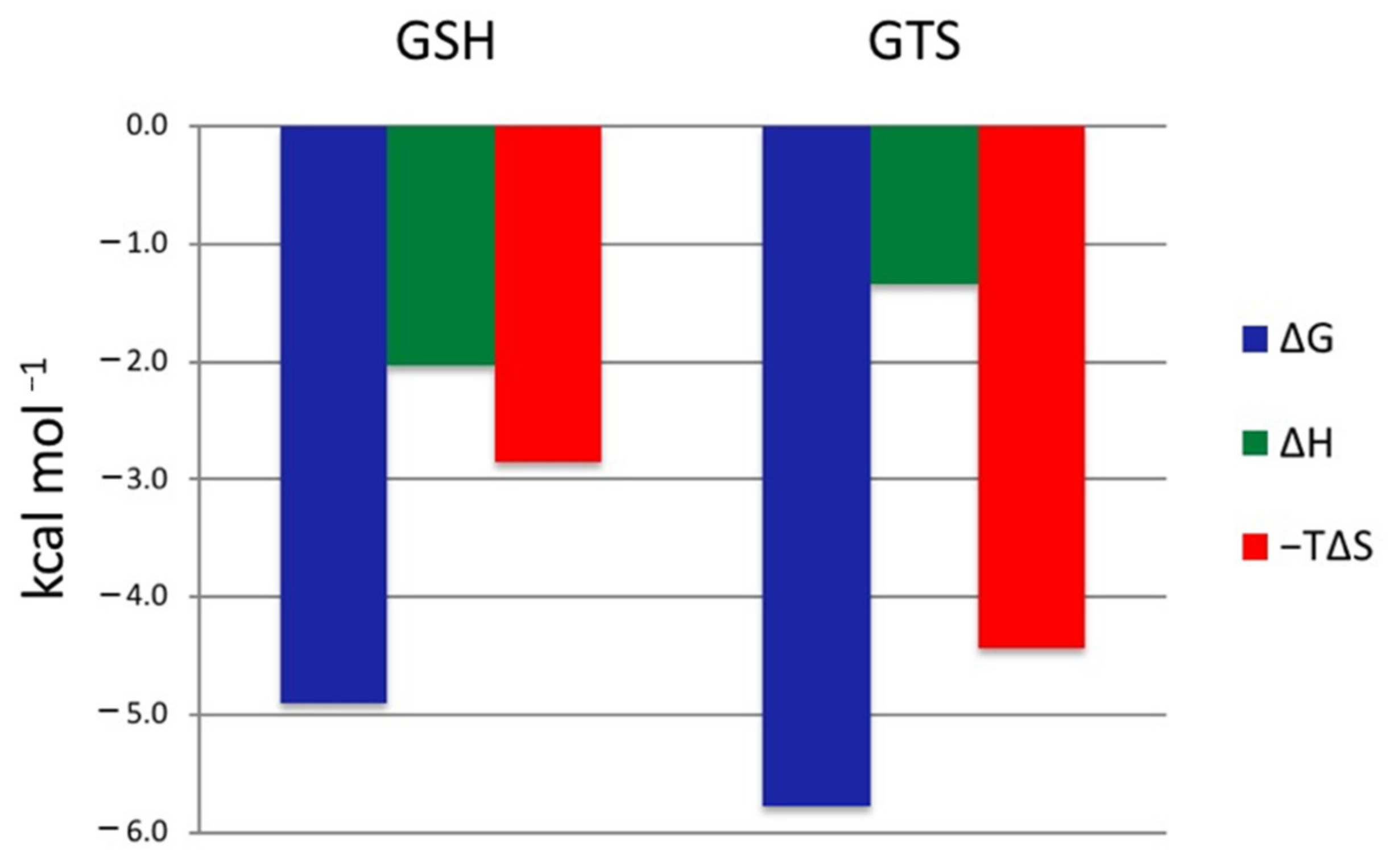
2.5. GSH and GTS Binding Thermodynamics
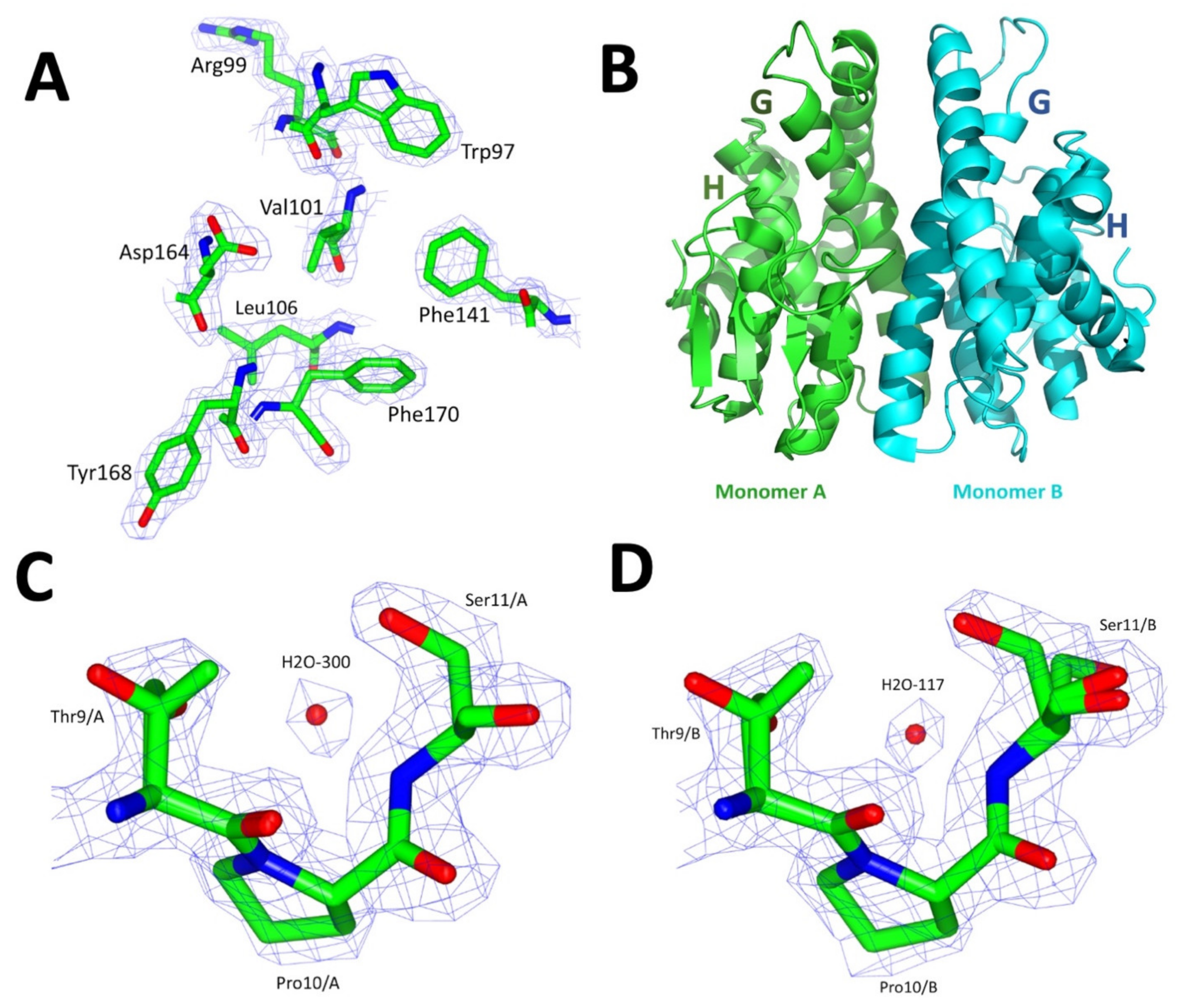
2.6. Crystallographic Structure of VpGSTT2
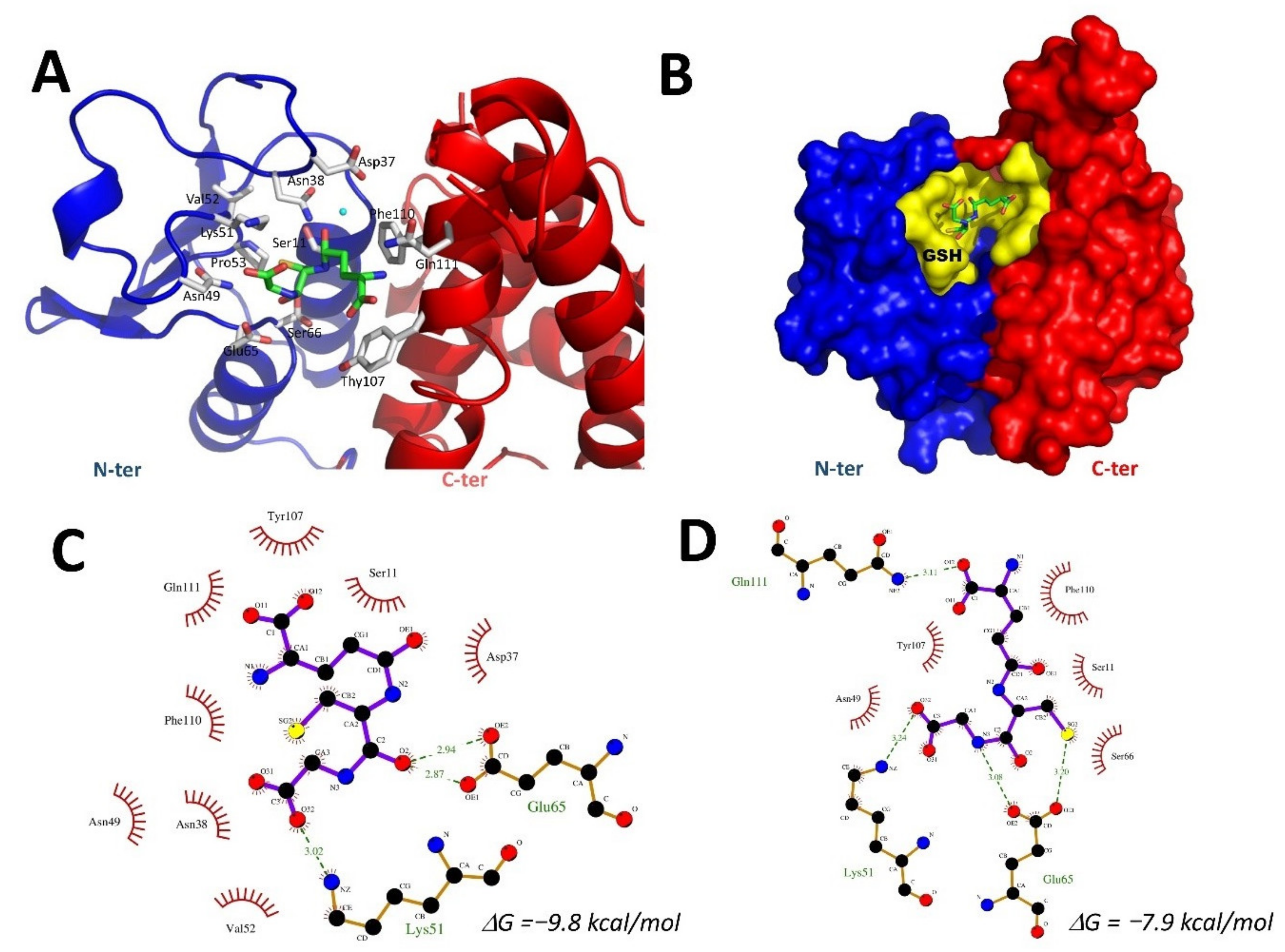
2.7. Docking of GSH into the VpGSTT2 Crystal Structure
3. Discussion
4. Materials and Methods
4.1. Overexpression and Purification of VpGSTT2
4.2. His-Tag Removal
4.3. Enzymatic Activity and Kinetic Parameters
4.4. Solvent Kinetic Isotope Effect: Proton Inventory
4.5. Isothermal Titration Calorimetry
4.6. Structure Determination
4.7. Molecular Docking
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Makino, K.; Oshima, K.; Kurokawa, K.; Yokoyama, K.; Uda, T.; Tagomori, K.; Iijima, Y.; Najima, M.; Nakano, M.; Yamashita, A.; et al. Genome sequence of Vibrio parahaemolyticus: A pathogenic mechanism distinct from that of V cholerae. Lancet 2003, 361, 743–749. [Google Scholar] [CrossRef]
- Caburlotto, G.; Gennari, M.; Ghidini, V.; Tafi, M.; Lleo, M.M. Serological and molecular characterization of Vibrio parahaemolyticus marine strains carrying pandemic genetic markers. ISME J. 2010, 4, 1071–1074. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Jimenez, S.; Noriega-Orozco, L.; Sotelo-Mundo, R.R.; Cantu-Robles, V.A.; Cobian-Guemes, A.G.; Cota-Verdugo, R.G.; Gamez-Alejo, L.A.; del Pozo-Yauner, L.; Guevara-Hernandez, E.; Garcia-Orozco, K.D.; et al. High-quality draft genomes of two Vibrio parahaemolyticus strains aid in understanding acute hepatopancreatic necrosis disease of cultured shrimps in Mexico. Genome Announc. 2014, 2, e00800-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waisberg, M.; Joseph, P.; Hale, B.; Beyersmann, D. Molecular and cellular mechanisms of cadmium carcinogenesis. Toxicology 2003, 192, 95–117. [Google Scholar] [CrossRef]
- Ma, X.-X.; Jiang, Y.; He, Y.-X.; Bao, R.; Chen, Y.; Zhou, C.-Z. Structures of yeast glutathione-S-transferase Gtt2 reveal a new catalytic type of GST family. EMBO Rep. 2009, 10, 1320–1326. [Google Scholar] [CrossRef]
- Choi, J.H.; Lou, W.; Vancura, A. A novel membrane-bound glutathione S-transferase functions in the stationary phase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 29915–29922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morel, M.; Ngadin, A.A.; Droux, M.; Jacquot, J.-P.; Gelhaye, E. The fungal glutathione S-transferase system. Evidence of new classes in the wood-degrading basidiomycete Phanerochaete chrysosporium. Cell. Mol. Life Sci. 2009, 66, 3711–3725. [Google Scholar] [CrossRef]
- Armstrong, R.N. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem. Res. Toxicol. 1997, 10, 2–18. [Google Scholar] [CrossRef]
- Dixon, D.; Sellars, J.; Edwards, R.H. The Arabidopsis phi class glutathione transferase AtGSTF2: Binding and regulation by biologically active heterocyclic ligands. Biochem. J. 2011, 438, 63–70. [Google Scholar] [CrossRef] [Green Version]
- Allocati, N.; Federici, L.; Masulli, M.; di Ilio, C. Glutathione transferases in bacteria. FEBS J. 2008, 276, 58–75. [Google Scholar] [CrossRef]
- Allocati, N.; Casalone, E.; Masulli, M.; Polekhina, G.; Rossjohn, J.; Parker, M.W.; di Ilio, C. Evaluation of the role of two conserved active-site residues in Beta class glutathione S-transferases. Biochem. J. 2000, 351, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, D.; Meade, G.; Foley, V.M.; Dowd, C.A. Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001, 360, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Stourman, N.V.; Rose, J.H.; Vuilleumier, S.; Armstrong, R.N. Catalytic mechanism of dichloromethane dehalogenase from Methylophilus sp. strain DM11. Biochemistry 2003, 42, 11048–11056. [Google Scholar] [CrossRef] [PubMed]
- Anandarajah, K.; Kiefer, P.M.; Donohoe, B.S.; Copley, S.D. Recruitment of a double bond isomerase to serve as a reductive dehalogenase during biodegradation of pentachlorophenol. Biochemistry 2000, 39, 5303–5311. [Google Scholar] [CrossRef] [PubMed]
- Wadington, M.C.; Ladner, J.E.; Stourman, N.V.; Harp, J.M.; Armstrong, R.N. Analysis of the structure and function of YfcG from Escherichia coli reveals an efficient and unique disulfide bond reductase. Biochemistry 2009, 48, 6559–6561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burns, C.; Geraghty, R.; Neville, C.; Murphy, A.; Kavanagh, K.; Doyle, S. Identification, cloning, and functional expression of three glutathione transferase genes from Aspergillus fumigatus. Fungal Genet. Biol. 2005, 42, 319–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014, 2014, 1–31. [Google Scholar] [CrossRef]
- Contreras-Vergara, C.A.; Valenzuela-Soto, E.; García-Orozco, K.D.; Sotelo-Mundo, R.; Yepiz-Plascencia, G. A Mu-class glutathione S-transferase from gills of the marine shrimp Litopenaeus vannamei: Purification and characterization. J. Biochem. Mol. Toxicol. 2007, 21, 62–67. [Google Scholar] [CrossRef]
- Arca, P.; Hardisson, C.; Suárez, J. Purification of a glutathione S-transferase that mediates fosfomycin resistance in bacteria. Antimicrob. Agents Chemother. 1990, 34, 844–848. [Google Scholar] [CrossRef] [Green Version]
- Roxas, V.P.; Smith, R.K.; Allen, E.R.; Allen, R.D. Overexpression of glutathione S-transferase/glutathioneperoxidase enhances the growth of transgenic tobacco seedlings during stress. Nat. Biotechnol. 1997, 15, 988–991. [Google Scholar] [CrossRef]
- Valenzuela-Chavira, I.; Contreras-Vergara, C.A.; Arvizu-Flores, A.; Serrano-Posada, H.; Lopez-Zavala, A.A.; García-Orozco, K.D.; Hernandez-Paredes, J.; Rudino-Pinera, E.; Stojanoff, V.; Sotelo-Mundo, R.R.; et al. Insights into ligand binding to a glutathione S-transferase from mango: Structure, thermodynamics and kinetics. Biochimie 2017, 135, 35–45. [Google Scholar] [CrossRef] [Green Version]
- McNicholas, S.; Potterton, E.; Wilson, K.; Noble, M. Presenting your structures: TheCCP4mgmolecular-graphics software. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 386–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chronopoulou, E.; Madesis, P.; Asimakopoulou, B.; Platis, D.; Tsaftaris, A.; Labrou, N.E. Catalytic and structural diversity of the fluazifop-inducible glutathione transferases from Phaseolus vulgaris. Planta 2012, 235, 1253–1269. [Google Scholar] [CrossRef] [Green Version]
- Rhee, J.-S.; Lee, Y.-M.; Hwang, D.-S.; Won, E.-J.; Raisuddin, S.; Shin, K.-H.; Lee, J.-S. Molecular cloning, expression, biochemical characteristics, and biomarker potential of theta class glutathione S-transferase (GST-T) from the polychaete Neanthes succinea. Aquat. Toxicol. 2007, 83, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Vontas, J.G.; Small, G.J.; Nikou, D.C.; Ranson, H.; Hemingway, J. Purification, molecular cloning and heterologous expression of a glutathione S-transferase involved in insecticide resistance from the rice brown planthopper, Nilaparvata lugens. Biochem. J. 2002, 362, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Ranson, H.; Prapanthadara, L.-A.; Hemingway, J. Cloning and characterization of two glutathione S-transferases from a DDT-resistant strain of Anopheles gambiae. Biochem. J. 1997, 324, 97–102. [Google Scholar] [CrossRef] [Green Version]
- Tang, A.; Tu, C.P. Biochemical characterization of Drosophila glutathione S-transferases D1 and D21. J. Biol. Chem. 1994, 269, 27876–27884. [Google Scholar] [CrossRef]
- Board, P.G.; Coggan, M.; Wilce, M.C.J.; Parker, M. Evidence for an essential serine residue in the active site of the Theta class glutathione transferases. Biochem. J. 1995, 311, 247–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Axarli, I.; Dhavala, P.; Papageorgiou, A.; Labrou, N. Crystallographic and functional characterization of the fluorodifen-inducible glutathione transferase from Glycine max reveals an active site topography suited for diphenylether herbicides and a novel L-site. J. Mol. Biol. 2009, 385, 984–1002. [Google Scholar] [CrossRef]
- Skopelitou, K.; Muleta, A.W.; Papageorgiou, A.; Chronopoulou, E.; Labrou, N.E. Catalytic features and crystal structure of a tau class glutathione transferase from Glycine max specifically upregulated in response to soybean mosaic virus infections. Biochim. Biophys. Acta BBA Proteins Proteom. 2015, 1854, 166–177. [Google Scholar] [CrossRef] [Green Version]
- Dixon, D.; McEwen, A.G.; Lapthorn, A.; Edwards, R. Forced evolution of a herbicide detoxifying glutathione transferase. J. Biol. Chem. 2003, 278, 23930–23935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, T.; Yang, Z.-L.; Yang, X.; Liu, Y.-J.; Wang, X.-R.; Zeng, Q.-Y. Extensive functional diversification of the Populus glutathione S-transferase supergene family. Plant Cell 2010, 21, 3749–3766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piero, A.R.L.; Mercurio, V.; Puglisi, I.; Petrone, G. Different roles of functional residues in the hydrophobic binding site of two sweet orange tau glutathione S-transferases. FEBS J. 2009, 277, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Schowen, K.B.; Schowen, R.L. Solvent isotope effects on enzyme systems. In Methods in Enzymology; Elsevier BV: Amsterdam, The Netherlands, 1982; Volume 87, pp. 551–606. [Google Scholar]
- Quinn, D.M.; Sutton, L.D. Theoretical basis and mechanistic utility of solvent isotope effects. In Enzyme Mechanism from Isotope Effects; Cook, P.F., Ed.; CRC Press: Boca Raton, FL, USA, 1991; pp. 73–126. [Google Scholar]
- Takeda, M.; Jee, J.; Terauchi, T.; Kainosho, M. Detection of the sulfhydryl groups in proteins with slow hydrogen exchange rates and determination of their proton/deuteron fractionation factors using the deuterium-induced effects on the 13Cβ NMR signals. J. Am. Chem. Soc. 2010, 132, 6254–6260. [Google Scholar] [CrossRef]
- Weiss, P.M.; Cook, P.F.; Hermes, J.D.; Cleland, W.W. Evidence from nitrogen-15 and solvent deuterium isotope effects on the chemical mechanism of adenosine deaminase. Biochemistry 1987, 26, 7378–7384. [Google Scholar] [CrossRef]
- Moynihan, M.M.; Murkin, A.S. Cysteine is the general base that serves in catalysis by isocitrate lyase and in mecha-nism-based inhibition by 3-nitropropionate. Biochemistry 2013, 53, 178–187. [Google Scholar] [CrossRef]
- Susan-Resiga, D.; Nowak, T. The proton transfer step catalyzed by yeast pyruvate kinase. J. Biol. Chem. 2003, 278, 12660–12671. [Google Scholar] [CrossRef] [Green Version]
- Parsons, J.F.; Armstrong, R.N. Proton configuration in the ground state and transition state of a glutathione transfer-ase-catalyzed reaction inferred from the properties of tetradeca (3-fluorotyrosyl) glutathione transferase. J. Am. Chem. Soc. 1996, 118, 2295–2296. [Google Scholar] [CrossRef]
- Ortiz-Salmeron, E.; Nuccetelli, M.; Oakley, A.; Parker, M.; Bello, M.L.; Fuentes, L.S.G. Thermodynamic description of the effect of the mutation Y49F on human glutathione transferase P1-1 in binding with glutathione and the inhibitor S-hexylglutathione. J. Biol. Chem. 2003, 278, 46938–46948. [Google Scholar] [CrossRef] [Green Version]
- Quesada-Soriano, I.; Barón, C.; García-Maroto, F.; Aguilera, A.M.; García-Fuentes, L. Calorimetric studies of ligands binding to glutathione S-transferase from the malarial parasite Plasmodium falciparum. Biochemistry 2013, 52, 1980–1989. [Google Scholar] [CrossRef]
- Holm, L. DALI and the persistence of protein shape. Protein Sci. 2020, 29, 128–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerlt, J.A.; Allen, K.N.; Almo, S.C.; Armstrong, R.N.; Babbitt, P.C.; Cronan, J.E.; Dunaway-Mariano, D.; Imker, H.J.; Jacobson, M.P.; Minor, W.; et al. The enzyme function initiative. Biochemistry 2011, 50, 9950–9962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 3, W270–W277. [Google Scholar] [CrossRef] [Green Version]
- Ochoa-Leyva, A.; Soberón, X.; Sánchez, F.; Argüello, M.; Montero-Morán, G.; Saab-Rincón, G. Protein design through systematic catalytic loop exchange in the (β/α)8 fold. J. Mol. Biol. 2009, 387, 949–964. [Google Scholar] [CrossRef]
- Hood, L.E.; Aebersold, R. Foreword. Curr. Protoc. Protein Sci. 1997, 7, i. Available online: https://ur.booksc.eu/book/64802026/241c9f (accessed on 23 August 2021). [CrossRef]
- Brunelle, J.L.; Green, R. One-dimensional SDS-polyacrylamide gel electrophoresis (1D SDS-PAGE). Methods Enzymol. 2014, 541, 151–159. [Google Scholar] [CrossRef]
- Ladner-Keay, C.L.; Turner, R.J.; Edwards, R.A. Fluorescent Protein Visualization Immediately After Gel Electrophoresis Using an In-Gel Trichloroethanol Photoreaction with Tryptophan; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2018; pp. 179–190. [Google Scholar]
- López-Zavala, A.A.; Garcia-Orozco, K.D.; Carrasco-Miranda, J.S.; Sugich-Miranda, R.; Velazquez-Contreras, E.F.; Criscitiello, M.F.; Brieba, L.G.; Rudiño-Piñera, E.; Sotelo-Mundo, R.R. Crystal structure of shrimp arginine kinase in binary complex with arginine—A molecular view of the phosphagen precursor binding to the enzyme. J. Bioenerg. Biomembr. 2013, 45, 511–518. [Google Scholar] [CrossRef]
- Carrasco-Miranda, J.S.; Lopez-Zavala, A.A.; Arvizu-Flores, A.A.; Garcia-Orozco, K.D.; Stojanoff, V.; Rudiño-Piñera, E.; Brieba, L.G.; Sotelo-Mundo, R.R. Crystal structure of the shrimp proliferating cell nuclear antigen: Structural complementarity with WSSV DNA polymerase PIP-Box. PLoS ONE 2014, 9, e94369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habig, W.H.; Pabst, M.J.; Jakoby, W.B. Glutathione S-transferases: The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974, 249, 7130–7139. [Google Scholar] [CrossRef]
- Jencks, W.P. Catalysis in Chemistry and Enzimology; Dover Publications, Inc.: New York, NY, USA, 1987. [Google Scholar]
- Leskovac, V. Comprehensive Enzyme Kinetics; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2003. [Google Scholar]
- Schowen, R.L. The use of solvent isotope effects in the pursuit of enzyme mechanisms. J. Label. Compd. Radiopharm. 2007, 50, 1052–1062. [Google Scholar] [CrossRef]
- Schowen, K.; Limbach, H.-H.; Denisov, G.; Schowen, R. Hydrogen bonds and proton transfer in general-catalytic transition-state stabilization in enzyme catalysis. Biochim. Biophys. Acta BBA Bioenerg. 2000, 1458, 43–62. [Google Scholar] [CrossRef] [Green Version]
- Dumas, P.; Ennifar, E.; Bec, G.; Piñeiro, A.; Sabn, J.; Muñoz, E.; Rial, J.; Emprendia, C.V.; Santiago de Compostela, A. Implementation of KinITC into AFFINImeter; Malvern Instruments Limited: Worcestershire, UK, 2015. [Google Scholar]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of theCCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Heras, B.; Martin, J.L. Post-crystallization treatments for improving diffraction quality of protein crystals. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005, 61, 1173–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabsch, W. Research papers XDS research papers. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collaborative, C.P. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 1994, 50, 760. [Google Scholar]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005, 62, 72–82. [Google Scholar] [CrossRef]
- Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.; Winn, M.D.; Storoni, L.C.; Read, R. Phasercrystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Terwilliger, T.C.; Grosse-Kunstleve, R.W.; Afonine, P.V.; Moriarty, N.W.; Zwart, P.H.; Hung, L.-W.; Read, R.; Adams, P.D. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007, 64, 61–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, V.B.; Arendall, W.B., III; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem substance and compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- DeLano, W.L. The PyMOL Molecular Graphics System. Available online: https://github.com/schrodinger/pymol-open-source (accessed on 23 August 2021).
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- Boyd, C. Environmental factors and acute hepatopancreatic necrosis disease (AHPND) in shrimp ponds in Viet Nam: Practices for reducing risks. Asian Fish. Sci. 2018, 31S, 121–136. [Google Scholar] [CrossRef]
- Khanna, K.; Jamwal, V.L.; Kohli, S.K.; Gandhi, S.G.; Ohri, P.; Bhardwaj, R.; Abd_Allah, E.; Hashem, A.; Ahmad, P. Plant growth promoting rhizobacteria induced Cd tolerance in Lycopersicon esculentum through altered antioxidative defense expression. Chemosphere 2019, 217, 463–474. [Google Scholar] [CrossRef]
- Dourado, M.N.; Souza, L.A.; Martins, P.F.; Peters, L.P.; Piotto, F.; Azevedo, R.A. Burkholderia sp. SCMS54 triggers a global stress defense in tomato enhancing cadmium tolerance. Water Air Soil Pollut. 2014, 225, 1–16. [Google Scholar] [CrossRef]
- Dourado, M.N.; Franco, M.R.; Peters, L.P.; Martins, P.F.; Souza, L.A.; Piotto, F.; Azevedo, R.A. Antioxidant enzymes activities of Burkholderia spp. strains—Oxidative responses to Ni toxicity. Environ. Sci. Pollut. Res. 2015, 22, 19922–19932. [Google Scholar] [CrossRef]
- Lee, C.-T.; Chen, I.-T.; Yang, Y.-T.; Ko, T.-P.; Huang, Y.-T.; Huang, J.-Y.; Huang, M.-F.; Lin, S.-J.; Chen, C.-Y.; Lin, S.-S.; et al. The opportunistic marine pathogen Vibrio parahaemolyticusbecomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc. Natl. Acad. Sci. USA 2015, 112, 10798–10803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barboza, G.; Gorlach-Lira, K.; Sassi, C.; Sassi, R. Microcystins production and antibacterial activity of cyanobacterial strains of Synechocystis, Synechococcus and Romeria isolated from water and coral reef organisms of Brazilian coast. Rev. Biol. Trop. 2017, 65, 890. [Google Scholar] [CrossRef] [Green Version]
- Long, R.A.; Rowley, D.C.; Zamora, E.; Liu, J.; Bartlett, D.H.; Azam, F. Antagonistic interactions among marine bacteria impede the proliferation of Vibrio cholerae. Appl. Environ. Microbiol. 2005, 71, 8531–8536. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| GST | Specific Activity (units/mg) | Reference |
|---|---|---|
| VpGSTT2 | 5.66 | This work |
| VpGSTT2 S11A | 0.27 | This work |
| VpGSTT2 T9A/S11A | not detectable | This work |
| Human liver | 304–1297 | [17] |
| Aedes aegiptis Epsilon-class | 2.7–20.8 | [22] |
| Lipopenaeus vannamei Mu-class | 440.12 | [18] |
| Aspergillus fumigatus | 0.025 | [16] |
| Nicotiana tabacum | 48.61 | [20] |
| Mangifera indicaSaccharomyces cerevisiae GTT2 | 43.76 0.63 | [21] [5] |
| Escherichia coli | 0.52 | [19] |
| GSH | CDNB | ||||||
|---|---|---|---|---|---|---|---|
| Enzyme | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) | Reference |
| VpGSTT2 | 2.60 | 8.7 | 3.3 | 1.00 | 49 | 49 | This work |
| NlGST1-1 | 0.66 | 118 | 180 | 0.26 | 118 | 447 | [25] |
| AgGST1-1 | 0.807 | 97.3 | 120.7 | 0.12 | 97.4 | 792 | [26] |
| DmGST1-1 | 0.28 | 28.3 | 101 | 0.80 | 38.3 | 48 | [27] |
| LcGSTT | 0.51 | 11.8 | 23.2 | 0.15 | 9 | 60.1 | [28] |
| NsGSTT | 2.50 | 0.013 | ND | 3.11 | 0.015 | ND | [24] |
| GmGSTU4 | 0.159 | 6.05 | 38 | 0.16 | 2.48 | 15.7 | [29] |
| GmGSTU10 | 0.068 | 2.65 | 39 | 0.28 | 2.66 | 9.5 | [30] |
| MiGSTU | 0.69 | 89.52 | 129 | 0.79 | 68.49 | 86.51 | [21] |
| ZmGSTU1 | 0.56 | NR | NR | 1.01 | 18.6 | 18.4 | [31] |
| ZmGSTU2 | 1.72 | NR | NR | 0.12 | 34.5 | 300 | [31] |
| PtGSTU22 | 0.56 | 1118.39 | 1997.12 | 1.72 | 574.6 | 334.07 | [32] |
| PvGSTU2-2 | 0.05 | 10.84 | 217 | 0.86 | 21.2 | 24.7 | [23] |
| PvGSTU1-1 | 0.17 | 0.08 | 0.47 | ND | ND | ND | [23] |
| CsGSTU1 | 0.5 | 0.014 | 0.028 | 0.75 | 0.024 | 0.032 | [33] |
| CsGSTU2 | 0.5 | 0.077 | 0.154 | 1.0 | 0.108 | 0.108 | [33] |
| GST | Kd (μM) | ΔG (kcal/mol) | ΔH (kcal/mol) | −TΔS (kcal/mol) | Reference |
|---|---|---|---|---|---|
| VpGSTT2-GSH | 257.8 | −4.9 | −2.00 | −2.9 | This work |
| VpGSTT2-GTS | 57.9 | −5.8 | −1.4 | −4.4 | This work |
| MiGSTU-GSH | 5.2 | −7.2 | −26.4 | 19.2 | [21] |
| MiGSTU-GSX | 7.8 | −6.9 | −6.2 | −0.71 | [21] |
| hGSTP1-GSH | 85.98 | −5.53 | −11.21 | 5.65 | [41] |
| hGSTP1-GSX | 1.23 | −8.04 | −16.13 | 8.04 | [41] |
| SjGST-GSH | 0.99 | −4.9 | −5.7 | 0.8 | [41] |
| AtGST-GSX | 22.7 | −26.1 | −5.2 | −20.9 | [9] |
| PfGST-GSH | 140 | −5.7 | −11.8 | 6.1 | [42] |
| PfGST-GTS | 162 | −7.1 | −12.8 | 5.7 | [42] |
| Parameters | VpGSTT2-Apo |
|---|---|
| Data Collection Statics | |
| Space Group | P21 |
| Unit cell dimensions | |
| a, b, c (Å) | 56.3 50.4 69.6 |
| α, β, γ (degrees) | 90.0 90.1 90.0 |
| Resolution range (Å) | 40.8–1.92 |
| No. of reflections | 152,812 (20,045) |
| No. of unique reflections | 29,139 (3971) |
| Completeness (%) | 98.8% (94.3) |
| Rmeas (%) | 5.4 % (18) |
| CC1/2 (%) | 99.8 % (94.3) |
| I/σ(I) | 5.8 (2.1) |
| Multiplicity | 5.2 (5.0) |
| Monomers per asymmetric unit | 2 |
| Refinement statistics | |
| Resolution range (Å) | 40.8–1.92 |
| Rwork/Rfree (%) | 16.3/20.8 |
| Number of reflections | 29,106 (2668) |
| Clash score | 5.75 |
| Mean B-values (Å2) | |
| Protein | 28.6 |
| Ion/Ligand | 36.5 |
| Water | 33.6 |
| All atoms | 29.1 |
| Wilson plot | 23.4 |
| RMSD from ideal stereochemistry | |
| Bond lengths (Å) | 0.01 |
| Bond angles (degrees) | 0.61 |
| Coordinate error (Maximum-Likelihood Base) | 0.21 |
| Ramachandran plot (%) | |
| Most favored region | 96.3 |
| Additional allowed regions | 2.3 |
| Disallowed regions | 1.4 |
| PDB code | 7MIQ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valenzuela-Chavira, I.; Corona-Martinez, D.O.; Garcia-Orozco, K.D.; Beltran-Torres, M.; Sanchez-Lopez, F.; Arvizu-Flores, A.A.; Sugich-Miranda, R.; Lopez-Zavala, A.A.; Robles-Zepeda, R.E.; Islas-Osuna, M.A.; et al. A Novel Glutathione S-Transferase Gtt2 Class (VpGSTT2) Is Found in the Genome of the AHPND/EMS Vibrio parahaemolyticus Shrimp Pathogen. Toxins 2021, 13, 664. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13090664
Valenzuela-Chavira I, Corona-Martinez DO, Garcia-Orozco KD, Beltran-Torres M, Sanchez-Lopez F, Arvizu-Flores AA, Sugich-Miranda R, Lopez-Zavala AA, Robles-Zepeda RE, Islas-Osuna MA, et al. A Novel Glutathione S-Transferase Gtt2 Class (VpGSTT2) Is Found in the Genome of the AHPND/EMS Vibrio parahaemolyticus Shrimp Pathogen. Toxins. 2021; 13(9):664. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13090664
Chicago/Turabian StyleValenzuela-Chavira, Ignacio, David O. Corona-Martinez, Karina D. Garcia-Orozco, Melissa Beltran-Torres, Filiberto Sanchez-Lopez, Aldo A. Arvizu-Flores, Rocio Sugich-Miranda, Alonso A. Lopez-Zavala, Ramon E. Robles-Zepeda, Maria A. Islas-Osuna, and et al. 2021. "A Novel Glutathione S-Transferase Gtt2 Class (VpGSTT2) Is Found in the Genome of the AHPND/EMS Vibrio parahaemolyticus Shrimp Pathogen" Toxins 13, no. 9: 664. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13090664






