Pasteurella multocida Toxin Activates Various Heterotrimeric G Proteins by Deamidation
Abstract
:1. Introduction
2. Signal Transduction and Molecular Mechanism of PMT

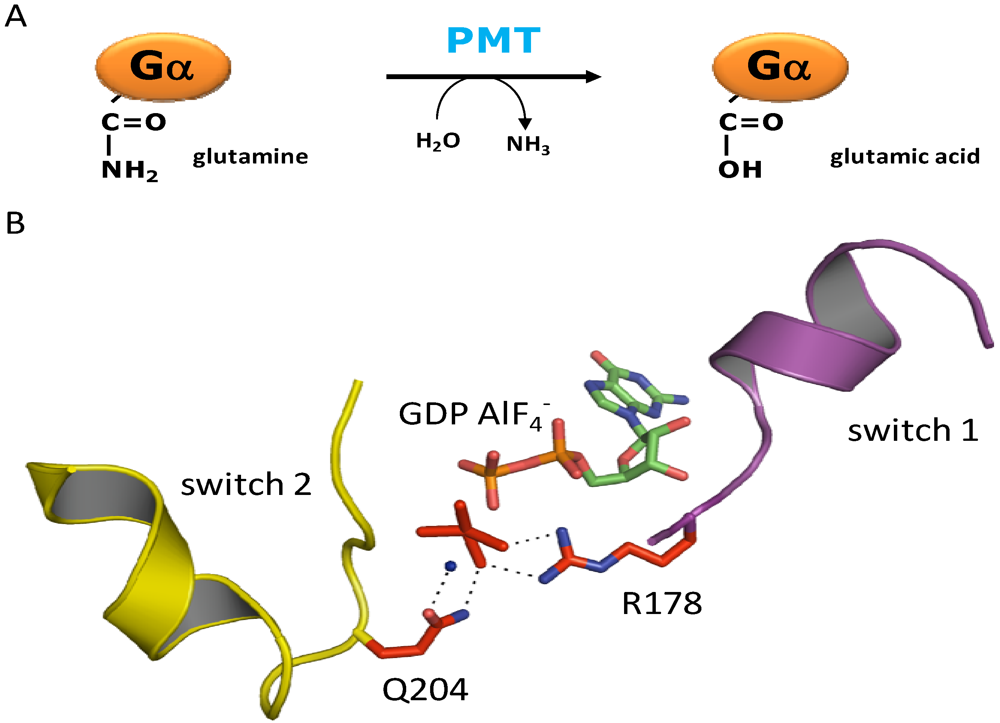
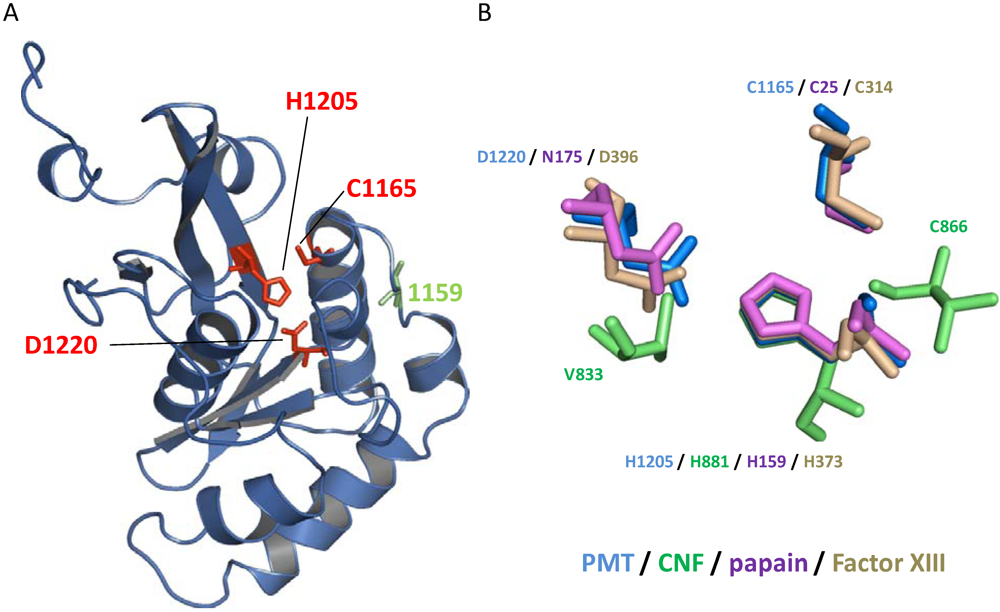
3. Structure

4. Toxin Uptake
5. Conclusions
Acknowledgements
References
- Kristinsson, G. Pasteurella multocida infections. Pediatr. Rev. 2007, 28, 472–473. [Google Scholar] [PubMed]
- Franque, L.W. Was ist die Schnueffelkrankheit der Schweine? In Teutsche Zeitschrift fuer die gesammte Thierheilkunde; 1830; Volume 1, pp. 75–77. [Google Scholar]
- Dominick, M.A.; Rimler, R.B. Turbinate atrophy in gnotobiotic pigs intranasally inoculated with protein toxin isolated from type D Pasteurella multocida. Am. J. Vet. Res. 1986, 47, 1532–1536. [Google Scholar] [PubMed]
- Lax, A.J.; Chanter, N. Cloning of the toxin gene from Pasteurella multocida and its role in atrophic rhinitis. J. Gen. Microbiol. 1990, 136, 81–87. [Google Scholar] [PubMed]
- Rozengurt, E.; Higgins, T.; Chanter, N.; Lax, A.J.; Staddon, J.M. Pasteurella multocida toxin: Potent mitogen for cultured fibroblasts. Proc. Natl. Acad. Sci. USA 1990, 87, 123–127. [Google Scholar]
- Seo, B.; Choy, E.W.; Maudsley, W.E.; Miller, W.E.; Wilson, B.A.; Luttrell, L.M. Pasteurella multocida toxin stimulates mitogen-activated protein kinase via Gq/11-dependent transactivation of the epidermal growth factor receptor. J. Biol. Chem. 2000, 275, 2239–2245. [Google Scholar] [PubMed]
- Sabri, A.; Wilson, B.A.; Steinberg, S.F. Dual actions of the Galpha(q) agonist Pasteurella multocida toxin to promote cardiomyocyte hypertrophy and enhance apoptosis susceptibility. Circ. Res. 2002, 90, 850–857. [Google Scholar] [PubMed]
- Wilson, B.A.; Zhu, X.; Ho, M.; Lu, L. Pasteurella multocida toxin activates the inositol triphosphate signaling pathway in Xenopus oocytes via Gqa-coupled phospholipase C-b1. J. Biol. Chem. 1997, 272, 1268–1275. [Google Scholar] [PubMed]
- Zywietz, A.; Gohla, A.; Schmelz, M.; Schultz, G.; Offermanns, S. Pleiotropic effects of Pasteurella multocida toxin are mediated by Gq-dependent and -independent mechanisms. Involvement of Gq but not G11. J. Biol. Chem. 2001, 276, 3840–3845. [Google Scholar] [PubMed]
- Orth, J.H.; Lang, S.; Aktories, K. Action of Pasteurella multocida toxin depends on the helical domain of Galphaq. J. Biol. Chem. 2004, 279, 34150–34155. [Google Scholar] [PubMed]
- Orth, J.H.; Aktories, K.; Kubatzky, K.F. Modulation of host cell gene expression through activation of STAT transcription factors by Pasteurella multocida toxin. J. Biol. Chem. 2007, 282, 3050–3057. [Google Scholar] [PubMed]
- Kozasa, T.; Jiang, X.; Hart, M.J.; Sternweis, P.M.; Singer, W.D.; Gilman, A.G.; Bollag, G.; Sternweis, P.C. p115 RhoGEF, a GTPase activating protein for Galpha12 and Galpha13. Science 1998, 280, 2109–2111. [Google Scholar] [PubMed]
- Lutz, S.; Freichel-Blomquist, A.; Yang, Y.; Rumenapp, U.; Jakobs, K.H.; Schmidt, M.; Wieland, T. The guanine nucleotide exchange factor p63RhoGEF, a specific link between Gq/11-coupled receptor signaling and RhoA. J. Biol. Chem. 2005, 280, 11134–11139. [Google Scholar] [PubMed]
- Orth, J.H.; Lang, S.; Taniguchi, M.; Aktories, K. Pasteurella multocida toxin-induced activation of RhoA is mediated via two families of G{alpha} proteins, G{alpha}q and G{alpha}12/13. J. Biol. Chem. 2005, 280, 36701–36707. [Google Scholar] [PubMed]
- Orth, J.H.; Fester, I.; Preuss, I.; Agnoletto, L.; Wilson, B.A.; Aktories, K. Activation of Galphai and subsequent uncoupling of receptor-Galphai signaling by Pasteurella multocida toxin. J. Biol. Chem. 2008, 283, 23288–23294. [Google Scholar] [PubMed]
- Preuss, I.; Kurig, B.; Nurnberg, B.; Orth, J.H.; Aktories, K. Pasteurella multocida toxin activates Gbetagamma dimers of heterotrimeric G proteins. Cell. Signal. 2009, 21, 551–558. [Google Scholar] [PubMed]
- Orth, J.H.; Preuss, I.; Fester, I.; Schlosser, A.; Wilson, B.A.; Aktories, K. Pasteurella multocida toxin activation of heterotrimeric G proteins by deamidation. Proc. Natl. Acad. Sci. USA 2009, 106, 7179–7184. [Google Scholar]
- Tesmer, J.J.; Berman, D.M.; Gilman, A.G.; Sprang, S.R. Structure of RGS4 bound to AlF4--activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell 1997, 89, 251–261. [Google Scholar] [PubMed]
- Orth, J.H.; Lang, S.; Preuss, I.; Milligan, G.; Aktories, K. Action of Pasteurella multocida toxin on Galpha(q) is persistent and independent of interaction with G-protein-coupled receptors. Cell. Signal. 2007, 19, 2174–2182. [Google Scholar] [PubMed]
- Katada, T.; Oinuma, M.; Ui, M. Two guanine nucleotide-binding proteins in rat brain serving as the specific substrate of islet-activating protein, pertussis toxin. Interaction of the alpha-subunits with beta gamma-subunits in development of their biological activities. J. Biol. Chem. 1986, 261, 8182–8191. [Google Scholar] [PubMed]
- Petersen, S.K. The complete nucleotide sequence of the Pasteurella multocida toxin gene and evidence for a transcriptional repressor, TxaR. Mol. Microbiol. 1990, 4, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Lemichez, E.; Flatau, G.; Bruzzone, M.; Boquet, P.; Gauthier, M. Molecular localization of the Escherichia coli cytotoxic necrotizing factor CNF1 cell-binding and catalytic domains. Mol. Microbiol. 1997, 24, 1061–1070. [Google Scholar] [PubMed]
- Pullinger, G.D.; Sowdhamini, R.; Lax, A.J. Localization of functional domains of the mitogenic toxin of Pasteurella multocida. Infect. Immun. 2001, 69, 7839–7850. [Google Scholar] [PubMed]
- Nakai, T.; Kume, K. Purification of three fragments of the dermonecrotic toxin from Pasteurella multocida. Res Vet. Sci 1987, 42, 232–237. [Google Scholar] [PubMed]
- Wilson, B.A.; Ponferrada, V.G.; Vallance, J.E.; Ho, M.F. Localization of the intracellular activity domain of Pasteurella multocida toxin to the N terminus. Infect. Immunity 1999, 67, 80–87. [Google Scholar]
- Busch, C.; Orth, J.; Djouder, N.; Aktories, K. Biological activity of a C-terminal fragment of Pasteurella multocida toxin. Infect. Immun. 2001, 69, 3628–3634. [Google Scholar] [PubMed]
- Kitadokoro, K.; Kamitani, S.; Miyazawa, M.; Hanajima-Ozawa, M.; Fukui, A.; Miyake, M.; Horiguchi, Y. Crystal structures reveal a thiol protease-like catalytic triad in the C-terminal region of Pasteurella multocida toxin. Proc. Natl. Acad. Sci. USA 2007, 104, 5139–5144. [Google Scholar]
- Reinert, D.J.; Jank, T.; Aktories, K.; Schulz, G.E. Structural Basis for the Function of Clostridium difficile Toxin B. J. Mol. Biol. 2005, 351, 973–981. [Google Scholar] [PubMed]
- Aminova, L.R.; Luo, S.; Bannai, Y.; Ho, M.; Wilson, B.A. The C3 domain of Pasteurella multocida toxin is the minimal domain responsible for activation of Gq-dependent calcium and mitogenic signaling. Protein Sci 2008, 17, 1–5. [Google Scholar]
- Schmidt, G.; Sehr, P.; Wilm, M.; Selzer, J.; Mann, M.; Aktories, K. Gln63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor 1. Nature 1997, 387, 725–729. [Google Scholar] [PubMed]
- Flatau, G.; Lemichez, E.; Gauthier, M.; Chardin, P.; Paris, S.; Fiorentini, C.; Boquet, P. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature 1997, 387, 729–733. [Google Scholar] [PubMed]
- Horiguchi, Y.; Inoue, N.; Masuda, M.; Kashimoto, T.; Katahira, J.; Sugimoto, N.; Matsuda, M. Bordetella bronchiseptica dermonecrotizing toxin induces reorganization of actin stress fibers through deamidation of Gln-63 of the GTP-binding protein Rho. Proc. Natl. Acad. Sci. USA 1997, 94, 11623–11626. [Google Scholar]
- Masuda, M.; Betancourt, L.; Matsuzawa, T.; Kashimoto, T.; Takao, T.; Shimonishi, Y.; Horiguchi, Y. Activation of Rho through a cross-link with polyamines catalyzed by Bordetella dermonecrotizing toxin. EMBO J. 2000, 19, 521–530. [Google Scholar] [PubMed]
- Buetow, L.; Flatau, G.; Chiu, K.; Boquet, P.; Ghosh, P. Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1. Nature Struct. Biol. 2001, 8, 584–588. [Google Scholar]
- Pedersen, L.C.; Yee, V.C.; Bishop, P.D.; Trong, I.L.; Teller, D.C.; Stenkamp, R.E. Transglutaminase factor XIII uses proteinase-like catalytic triad to crosslink macromolecules. Protein Sci. 1994, 3, 1131–1135. [Google Scholar] [PubMed]
- Kashiwagi, T.; Yokoyama, K.; Ishikawa, K.; Ono, K.; Ejima, D.; Matsui, H.; Suzuki, E. Crystal structure of microbial transglutaminase from Streptoverticillium mobaraense. J. Biol. Chem. 2002, 277, 44252–44260. [Google Scholar] [PubMed]
- Pettit, R.K.; Ackermann, M.R.; Rimler, R.B. Receptor-mediated binding of Pasteurella multocida dermonecrotic toxin to canine osteosarcoma and monkey kidney (vero) cells. Labor. Invest. 1993, 69, 94–100. [Google Scholar]
- Staddon, J.M.; Barker, C.J.; Murphy, A.C.; Chanter, N.; Lax, A.J.; Michell, R.H.; Rozengurt, E. Pasteurella multocida toxin, a potent mitogen, increases inositol 1,4,5-triphosphate and mobilizes Ca2+ in swiss 3T3 cells. J. Biol. Chem. 1991, 266, 4840–4847. [Google Scholar] [PubMed]
- Baldwin, M.R.; Lakey, J.H.; Lax, A.J. Identification and characterization of the Pasteurella multocida toxin translocation domain. Mol. Microbiol. 2004, 54, 239–250. [Google Scholar] [PubMed]
- Van Raamsdonk, C.D.; Bezrookove, V.; Green, G.; Bauer, J.; Gaugler, L.; O'Brien, J.M.; Simpson, E.M.; Barsh, G.S.; Bastian, B.C. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 2008, 457, 599–602. [Google Scholar] [PubMed]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Orth, J.H.C.; Aktories, K. Pasteurella multocida Toxin Activates Various Heterotrimeric G Proteins by Deamidation. Toxins 2010, 2, 205-214. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins2020205
Orth JHC, Aktories K. Pasteurella multocida Toxin Activates Various Heterotrimeric G Proteins by Deamidation. Toxins. 2010; 2(2):205-214. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins2020205
Chicago/Turabian StyleOrth, Joachim H. C., and Klaus Aktories. 2010. "Pasteurella multocida Toxin Activates Various Heterotrimeric G Proteins by Deamidation" Toxins 2, no. 2: 205-214. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins2020205




