Biosynthesis and Toxicological Effects of Patulin
Abstract
:1. Introduction

2. Biosynthesis of Patulin
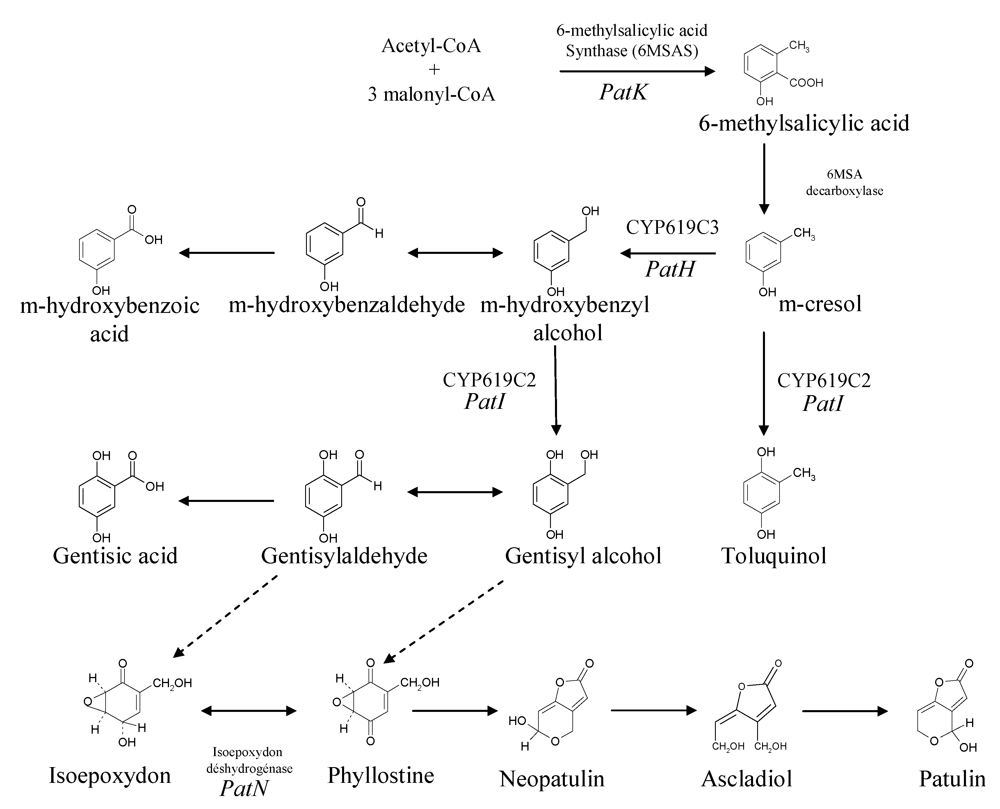
2.1. Precursors and enzymatic activities
2.2. Patulin gene cluster

2.3. Regulation of patulin biosynthesis
3. Toxicity of Patulin
3.1. General toxicity
3.2. Acute toxicity
3.3. Sub-acute toxicity
3.4. Genotoxicity
| Species | Dose | Duration | Observations | Reference |
|---|---|---|---|---|
| Mice | 24–36 mg/kg b.w. every day or every other day | 14 days | Intestinal disorders | [56] |
| Rat | 28–41 mg/kg b.w. every day or every other day | 14 days | Intestinal disorders | [58] |
| Rat | 25–295 mg/L in drinking water | 28 days | Decreased weight | [62] |
| Decreased Cl creatinine | ||||
| Gastric ulcers with high doses | ||||
| Rat | 0.1 mg/kg b.w. every day | 30 days | Decreased lipids | [65] |
| Decreased triglycerides | ||||
| Increased cholesterol | ||||
| Inhibition of intestinal ATPase | ||||
| Rat | 6–150 mg/L in drinking water | 13 weeks | Decreased food intake | [63] |
| Decreased weight with high doses | ||||
| Rat | 0.1 mg/kg b.w. every day | 60 & 90 days | Increased testosterone and LH levels | [67] |
| Alteration of testis and thyroid morphology | ||||
| Rat | 0.1 mg/kg b.w. every day | 60 & 90 days | Decreased sperm count | [68] |
| Alteration in sperm morphology | ||||
| Hamster | 16 mg/kg b.w. every day or every other day | 14 days | Intestinal disorders | [57] |
| Chicken | 100 µg every other day | 30 days | Intestinal disorders | [64] |
| Alteration of renal function | ||||
| Inhibition of intestinal and renal ATPases | ||||
| Monkey | 5; 50; 500 µg/kg b.w. then 5 mg/kg b.w. every day | 30 days 45 days | No toxicity | [66] |
| Food refusal (high dose) | ||||
| Alteration of renal function (medium dose) |
3.5. Cancerogenicity
3.6. Embryotoxicity and teratogenicity
3.7. Immunotoxicity
4. Conclusion
References and Notes
- Birkinshaw, J.H.; Michael, S.E.; Bracken, A.; Raistrick, H. Patulin in the common cold collaborative research on a derivative of Penicillium patulum Bainier. II. Biochemistry and Chemistry. Lancet 1943, 245, 625. [Google Scholar]
- Chalmers, I.; Clarke, M. Commentary: The 1944 patulin trial: The first properly controlled multicentre trial conducted under the aegis of the British Medical Research Council. Int. J. Epidemiol. 2004, 33, 253–260. [Google Scholar]
- European Union. Commission Regulation (EC) No. 1425/2003 of 11 August 2003 amending Regulation (EC) No 466/2001 as regards patulin. Official J. European Union L 203, 1–3.
- Moake, M.M.; Padilla-Zakour, O.I.; Worobo, R.W. Comprehensive review of patulin control methods in foods. Compr. Rev. Food Sci. Food Saf. 2005, 1, 8–21. [Google Scholar]
- Varga, J.; Due, M.; Frisvad, J.C.; Samson, R.A. Taxonomic revision of Aspergillus section Clavati based on molecular, morphological and physiological data. Stud. Mycol. 2007, 59, 89–106. [Google Scholar]
- Frisvad, J.C.; Smedsgaard, J.; Larsen, T.O.; Samson, R.A. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud. Mycol. 2004, 49, 201–242. [Google Scholar]
- Houbraken, J.; Samson, R.A.; Frisvad, J.C. Byssochlamys: Significance of heat resistance and mycotoxin production. Adv. Exp. Med. Biol. 2006, 571, 211–224. [Google Scholar]
- Puel, O.; Tadrist, S.; Delaforge, M.; Oswald, I.P.; Lebrihi, A. The inability of Byssochlamys fulva to produce patulin is related to absence of 6-methylsalicylic acid synthase and isoepoxydon dehydrogenase genes. Int. J. Food Microbiol. 2007, 115, 131–139. [Google Scholar]
- Samson, R.A.; Houbraken, J.; Varga, J.; Frisvad, J.C. Polyphasic taxonomy of the heat resistant ascomycete genus Byssochlamys and its Paecilomyces anamorphs. Persoonia 2009, 22, 14–27. [Google Scholar] [PubMed]
- McKinley, E.R.; Carlton, W.W. Patulin. In Mycotoxins and Phytoalexins; Sharma, R.P., Salunkhe, D.K., Eds.; CRC Press: Boca Raton, FL, USA, 1991; pp. 191–236. [Google Scholar]
- Birch, A.J.; Massy-Westrop, R.A.; Moye, C.J. Studies in relation to biosynthesis. VII. 2-Hydroxy-6-methylbenzoic acid in Penicillium griseofulvum Dierckx. Aust. J. Chem. 1955, 8, 539–544. [Google Scholar] [CrossRef]
- Lynen, F.; Tada, M. Die biochemischen grundlagen der polyacetat-regel. Angew. Chem. 1961, 73, 513–519. [Google Scholar]
- Sekiguchi, J.; Gaucher, G.M. Conidiogenesis and secondary metabolism in Penicillium urticae. Appl. Environ. Microbiol. 1977, 33, 147–158. [Google Scholar] [PubMed]
- Beck, J.; Ripka, S.; Siegner, A.; Schiltz, E.; Schweizer, E. The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum. Its gene structure relative to that of other polyketide synthases. Eur. J. Biochem. 1990, 192, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.K.; Reeves, C.; Gaucher, G.M. Isolation and sequencing of a genomic DNA clone containing the 3' terminus of the 6-methylsalicylic acid polyketide synthetase gene of Penicillium urticae. Can. J. Microbiol. 1991, 37, 86–95. [Google Scholar]
- Spencer, J.B.; Jordan, P.M. Purification and properties of 6-methylsalicylic acid synthase from Penicillium patulum. Biochem. J. 1992, 288, 839–846. [Google Scholar]
- Bu'Lock, J.D.; Ryan, A.J. The biogenesis of patulin. Proc. Chem. Soc. 1958, 222–223. [Google Scholar]
- Tanenbaum, S.W.; Bassett, E.W. The biosynthesis of patulin. III. Evidence for a molecular rearrangement of the aromatic ring. J. Biol. Chem. 1959, 234, 1861–1866. [Google Scholar] [PubMed]
- Forrester, P.I.; Gaucher, G.M. Conversion of 6-methylsalicylic acid into patulin by Penicillium urticae. Biochemistry 1972, 11, 1102–1107. [Google Scholar]
- Scott, A.I.; Zamir, L.; Phillips, G.T.; Yalpani, M. The biosynthesis of patulin. Bioorg. Chem. 1973, 2, 124–139. [Google Scholar]
- Scott, A.I.; Yalpani, M. A mass-spectrometric study of biosynthesis: Conversion of deuteron-m-cresol into patulin. Chem. Commun. 1967, 18, 945–946. [Google Scholar]
- Sekiguchi, J.; Gaucher, G.M. Isoepoxydon, a new metabolite of the patulin pathway in Penicillium urticae. Biochem. J. 1979, 182, 445–453. [Google Scholar]
- Sekiguchi, J.; Gaucher, G.M. Identification of phyllostine as an intermediate of the patulin pathway in Penicillium urticae. Biochemistry 1978, 17, 1785–1791. [Google Scholar]
- Sekiguchi, J.; Gaucher, G.M.; Yamada, Y. Biosynthesis of patulin in Penicillum urticae: Identification of isopatulin as a new intermediate. Tetrahedron Lett. 1979, 1, 41–42. [Google Scholar]
- Sekiguchi, J.; Shimamoto, T.; Yamada, Y.; Gaucher, G.M. Patulin biosynthesis: enzymatic and nonenzymatic transformations of the mycotoxin (E)-ascladiol. Appl. Environ. Microbiol. 1983, 45, 1939–1942. [Google Scholar]
- Priest, J.W; Light, R.J. Patulin biosynthesis: epoxidation of toluquinol and gentisyl alcohol by particulate preparations from Penicillium patulum. Biochemistry 1989, 28, 9192–9200. [Google Scholar] [CrossRef] [PubMed]
- Murphy, G.; Lynen, F. Patulin biosynthesis: The metabolism of m-hydroxybenzyl alcohol and m-hydroxybenzaldehyde by particulate preparations from Penicillium patulum. Eur. J. Biochem. 1975, 58, 467–475. [Google Scholar]
- Light, R.J. 6-methylsalicylic acid decarboxylase from Penicillium patulum. Biochim. Biophys. Acta 1969, 191, 430–438. [Google Scholar] [PubMed]
- Fedeshko, R.W. Polyketide enzymes and genes in Penicillium urticae. 1992. [Google Scholar]
- Keller, N.P.; Hohn, T.M. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 1997, 21, 17–29. [Google Scholar]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism - from biochemistry to genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar]
- Yu, J.; Chang, P.K.; Ehrlich, K.C.; Cary, J.W.; Bhatnagar, D.; Cleveland, T.E.; Payne, G.A.; Linz, J.E.; Woloshuk, C.P.; Bennett, J.W. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004, 70, 1253–1262. [Google Scholar] [PubMed]
- Seo, J.A.; Proctor, R.H.; Plattner, R.D. Characterization of four clustered and coregulated genes associated with fumonisin biosynthesis in Fusarium verticillioides. Fungal Genet. Biol. 2001, 34, 155–165. [Google Scholar]
- Kimura, M.; Tokai, T.; Takahashi-Ando, N.; Ohsato, S.; Fujimura, M. Molecular and genetic studies of Fusarium trichothecene biosynthesis: Pathways, genes, and evolution. Biosci. Biotechnol. Biochem. 2007, 71, 2105–2123. [Google Scholar]
- Kim, Y.T.; Lee, Y.R.; Jin, J.; Han, K.H.; Kim, H.; Kim, J.C.; Lee, T.; Yun, S.H.; Lee, Y.W. Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae. Mol. Microbiol. 2005, 58, 1102–1113. [Google Scholar]
- Dombrink-Kurtzman, M.A. The isoepoxydon dehydrogenase gene of the patulin metabolic pathway differs for Penicillium griseofulvum and Penicillium expansum. Antonie Van Leeuwenhoek 2006, 89, 1–8. [Google Scholar]
- Dombrink-Kurtzman, M.A. The sequence of the isoepoxydon dehydrogenase gene of the patulin biosynthetic pathway in Penicillium species. Antonie Van Leeuwenhoek 2007, 91, 179–189. [Google Scholar]
- White, S.; O'Callaghan, J.; Dobson, A.D. Cloning and molecular characterization of Penicillium expansum genes upregulated under conditions permissive for patulin biosynthesis. FEMS Microbiol.Lett. 2006, 255, 17–26. [Google Scholar]
- Dombrink-Kurtzman, M.A.; Engberg, A.E. Byssochlamys nivea with patulin-producing capability has an isoepoxydon dehydrogenase gene (idh) with sequence homology to Penicillium expansum and P. griseofulvum. Mycol. Res. 2006, 110, 1111–1118. [Google Scholar]
- Dombrink-Kurtzman, M.A. A gene having sequence homology to isoamyl alcohol oxidase is transcribed during patulin production in Penicillium griseofulvum. Curr. microbiol. 2008, 56, 224–228. [Google Scholar]
- Artigot, M.P.; Loiseau, N.; Laffitte, J.; Mas-Reguieg, L.; Tadrist, S.; Oswald, I.P.; Puel, O. Molecular cloning and functional characterization of two CYP619 cytochrome P450s involved in biosynthesis of patulin in Aspergillus clavatus. Microbiology 2009, 155, 1738–1747. [Google Scholar]
- Varga, J.; Tóth, B.; Kocsubé, S.; Farkas, B.; Szakács, G.; Téren, J.; Kozakiewicz, Z. Evolutionary relationships among Aspergillus terreus isolates and their relatives. Antonie Van Leeuwenhoek 2005, 88, 141–150. [Google Scholar]
- Fujii, I.; Ono, Y.; Tada, H.; Gomi, K.; Ebizuka, Y.; Sankawa, U. Cloning of the polyketide synthase gene atX from Aspergillus terreus and its identification as the 6-methylsalicylic acid synthase gene by heterologous expression. Mol. Gen. Genet. 1996, 253, 1–10. [Google Scholar]
- Read, G.; Westlake, D.W.; Vining, L.C. Quinone epoxides V The biosynthesis of terreic acid. Can. J. Biochem. 1969, 47, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Zhang, H. Transition metal-catalyzed nonoxidative decarboxylation reactions. Biochemistry 2006, 45, 10407–10411. [Google Scholar] [PubMed]
- Chanda, A.; Roze, L.V.; Kang, S.; Artymovich, K.A.; Hicks, G.R.; Raikhel, N.V.; Calvo, A.M.; Linz, J.E. A key role for vesicles in fungal secondary metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 19533–19538. [Google Scholar]
- Grootwassink, J.W.; Gaucher, G.M. De novo biosynthesis of secondary metabolism enzymes in homogeneous cultures of Penicillium urticae. J. Bacteriol. 1980, 141, 443–455. [Google Scholar]
- Rollins, M.J.; Gaucher, G.M. Ammonium repression of antibiotic and intracellular proteinase production in Penicillium urticae. Appl. Microbiol. Biotechnol. 1994, 41, 447–455. [Google Scholar]
- Feng, G.H.; Leonard, T.J. Culture conditions control expression of the genes for aflatoxin and sterigmatocystin biosynthesis in Aspergillus parasiticus and A. nidulans. Appl. Environ. Microbiol. 1998, 64, 2275–2277. [Google Scholar] [PubMed]
- Abbas, A.; Valez, H.; Dobson, A.D. Analysis of the effect of nutritional factors on OTA and OTB biosynthesis and polyketide synthase gene expression in Aspergillus ochraceus. Int. J. Food Microbiol. 2009, 135, 22–27. [Google Scholar] [PubMed]
- Summerer, S.M. A study of fungal polyketide gene expression and isolation. 1996. [Google Scholar]
- Ellis, C.M. Regulation of polyketide gene expression: The isolation and function of nitrogen regulatory factor, NRFA from Penicillium urticae. 1996. [Google Scholar]
- Scott, R.E.; Jones, A.; Lam, K.S.; Gaucher, G.M. Manganese and antibiotic biosynthesis. I. A specific manganese requirement for patulin production in Penicillium urticae. Can. J. Microbiol. 1986, 32, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.E.; Jones, A.; Gaucher, G.M. Manganese and antibiotic biosynthesis. III. The site of manganese control of patulin production in Penicillium urticae. Can. J. Microbiol. 1986, 32, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Gaucher, G.M.; Lam, K.S.; Grootwassink, J.W.D.; Neway, J.; Deo, Y.M. The initiation and longevity of patulin biosynthesis. Dev. Industr. Microbiol. 1981, 22, 219–232. [Google Scholar]
- McKinley, E.R.; Carlton, W.W. Patulin mycotoxicosis in Swiss ICR mice. Food Cosmet. Toxicol. 1980, 18, 181–187. [Google Scholar]
- McKinley, E.R.; Carlton, W.W. Patulin mycotoxicosis in the Syrian hamster. Food Cosmet. Toxicol. 1980, 18, 173–179. [Google Scholar]
- McKinley, E.R.; Carlton, W.W.; Boon, G.D. Patulin mycotoxicosis in the rat: Toxicology, pathology and clinical pathology. Food Chem. Toxicol. 1982, 20, 289–300. [Google Scholar]
- WHO. Joint FAO/WHO Expert Committee on Food additives (JECFA), Position paper on patulin, 30th session, 9–13 March, 1998.
- Hayes, A.W.; Phillips, T.D.; Williams, W.L.; Ciegler, A. Acute toxicity of patulin in mice and rats. Toxicology 1979, 13, 91–100. [Google Scholar]
- Ciegler, A.; Beckwith, A.C.; Jackson, L.K. Teratogenicity of patulin and patulin adducts formed with cysteine. Appl. Environ. Microbiol. 1976, 31, 664–667. [Google Scholar]
- Speijers, G.J.A.; Kolkman, R.; Franken, M.A.M.; Van Leeuwen, F.X.R.; Danse, L.H.J.C. Subacute toxiciteit van patulin in de rat. Rapport nr. 617903 001. Rijksinstituut voor Volksgezondheid en Milieuhygiëne, 1985. [Google Scholar]
- Speijers, G.J.A.; Franken, M.A.M.; Van Leeuwen, F.X.R.; Van Egmond, H.P.; Boot, R.; Loeber, J.G. Subchronic oral toxicity study of patulin in the rat. Report no. 618314 001. Rijksinstituut voor Volksgezondheid en Milieuhygiëne, 1986. [Google Scholar]
- Devaraj, H.; Suseela, R.E.; Devaraj, N. Patulin toxicosis in chicks. Curr. Sci. 1986, 55, 998–999. [Google Scholar]
- Devaraj, H.; Devaraj, N. Rat intestinal lipid changes in patulin toxicity. Indian J. of Exp. Biol. 1987, 25, 637–638. [Google Scholar]
- Garza, H.C.; Swanson, B.G.; Branen, A.L. Toxicology study of patulin in monkeys. J. Food Sci. 1977, 42, 1229–1231. [Google Scholar]
- Selmanoglu, G.; Kockaya, E.A. Investigation of the effects of patulin on thyroid and testis, and hormone levels in growing male rats. Food Chem. Toxicol. 2004, 42, 721–727. [Google Scholar]
- Selmanoglu, G. Evaluation of the reproductive toxicity of patulin in growing male rats. Food Chem. Toxicol. 2006, 44, 2019–2024. [Google Scholar]
- Wurgler, F.E.; Friederich, U.; Schlatter, J. Lack of mutagenicity of ochratoxin A and B, citrinin, patulin and cnestine in Salmonella typhimurium TAI02. Mutat. Res. 1991, 261, 209–216. [Google Scholar]
- IARC. Some Naturally Occurring and Synthetic Food Components, Furocoumarins and Ultraviolet Radiation. In Monographs on the Evaluation of the Carcinogenic Risk of Chemicals to Humans; IARC: Lyon, France, 1986; pp. 83–98. [Google Scholar]
- Sakai, M.; Abe, K.; Okumura, H.; Kawamura, O.; Sugiura, Y.; Horie, Y.; Ueno, Y. Genotoxicity of fungi evaluated by SOS microplate assay. Nat. Toxins 1992, 1, 27–34. [Google Scholar]
- Hradec, J.; Vesely, D. The initiator tRNA acceptance assay as a short-term test for carcinogens. Carcinogenesis 1989, 10, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Thust, R.; Kneist, S.; Mendel, J. Patulin, a further clastogenic mycotoxin, is negative in the SCE assay in Chinese hamster V79-E cells in vitro. Mutat. Res. 1982, 103, 91–97. [Google Scholar]
- Pfeiffer, E.; Gross, K.; Metzler, M. Aneuploidogenic and clastogenic potential of the mycotoxins citrinin and patulin. Carcinogenesis 1998, 19, 1313–1318. [Google Scholar]
- Alves, I.; Oliveira, N.G.; Laires, A.; Rodrigues, A.S.; Rueff, J. Induction of micronuclei and chromosomal aberrations by the mycotoxin patulin in mammalian cells: role of ascorbic acid as a modulator of patulin clastogenicity. Mutagenesis 2000, 15, 229–234. [Google Scholar]
- Umeda, M.; Tsutsui, T.; Saito, M. Mutagenicity and inducibility of DNA single-strand breaks and chromosome aberrations by various mycotoxins. Gann 1977, 68, 619–625. [Google Scholar]
- Schumacher, D.M.; Metzler, M.; Lehmann, L. Mutagenicity of the mycotoxin patulin in cultured Chinese hamster V79 cells, and its modulation by intracellular glutathione. Arch. Toxicol. 2005, 79, 110–121. [Google Scholar]
- Schumacher, D.M.; Wagner, J.; Metzler, M.; Lehmann, L. Influence of decreased intracellular glutathione level on the mutagenicity of patulin in cultured mouse lymphoma cells. Mycotoxin Res. 2005, 21, 150–152. [Google Scholar]
- Liu, B.H.; Yu, F.Y.; Wu, T.S.; Li, S.Y.; Su, M.C.; Wang, M.C.; Shih, S.M. Evaluation of genotoxic risk and oxidative DNA damage in mammalian cells exposed to mycotoxins, patulin and citrinin. Toxicol. Appl. Pharmacol. 2003, 191, 255–263. [Google Scholar]
- Liu, B.H.; Wu, T.S.; Yu, F.Y.; Su, C.C. Induction of oxidative stress response by the mycotoxin patulin in mammalian cells. Toxicol. Sci. 2007, 95, 340–347. [Google Scholar]
- Riley, R.T.; Showker, J.L. The mechanism of patulin’s cytotoxicity and the antioxidant activity of indole tetramic acids. Toxicol. Appl. Pharmacol. 1991, 109, 108–126. [Google Scholar]
- Osswald, H.; Frank, H.K.; Komitowski, D.; Winter, H. Long-term testing of patulin administered orally to Sprague-Dawley rats and Swiss mice. Food Cosmet. Toxicol. 1978, 16, 243–247. [Google Scholar]
- Becci, P.J.; Hess, F.G.; Johnson, W.D.; Gallo, M.A.; Babish, J.G.; Dailey, R.E.; Parent, R.A. Long-term carcinogenicity and toxicity studies of patulin in the rat. J. Appl. Toxicol. 1981, 1, 256–261. [Google Scholar]
- Dailey, R.E.; Brouwer, E.; Blaschka, A.M.; Reynaldo, E.F.; Green, S.; Monlux, W.S.; Ruggles, D.I. Intermediate-duration toxicity study of patulin in rats. J. Toxicol. Environ. Health 1977, 2, 713–725. [Google Scholar] [PubMed]
- Reddy, C.S.; Chan, P.K.; Hayes, A.W. Teratogenic and dominant lethal studies of patulin in mice. Toxicology 1978, 11, 219–223. [Google Scholar]
- Roll, R.; Matthiaschk, G.; Korte, A. Embryotoxicity and mutagenicity of mycotoxins. J. Environ. Pathol. Toxicol. Oncol. 1990, 10, 1–7. [Google Scholar]
- Smith, E.E.; Duffus, E.A.; Small, M.H. Effects of patulin on postimplantation rat embryos. Arch. Environ. Contam. Toxicol. 1993, 25, 267–270. [Google Scholar]
- Oswald, I.P.; Comera, C. Immunotoxicity of mycotoxins. Rev. Med. Vet. 1998, 149, 585–590. [Google Scholar]
- Sorenson, W.G.; Gerberick, G.F.; Lewis, D.M.; Castranova, V. Toxicity of mycotoxins for the rat pulmonary macrophage in vitro. Env. Health Persp. 1986, 66, 45–53. [Google Scholar]
- Bourdiol, D.; Escoula, L.; Salvayre, R. Effect of patulin on microbicidal activity of mouse peritoneal macrophages. Food Chem. Toxicol. 1990, 28, 29–33. [Google Scholar]
- Wichmann, G.; Herbarth, O.; Lehmann, I. The mycotoxins citrinin, gliotoxin, and patulin affect interferon-gamma rather than interleukin-4 production in human blood cells. Environ. Toxicol. 2002, 17, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Luft, P.; Oostingh, G.J.; Gruijthuijsen, Y.; Horejs-Hoeck, J.; Lehmann, I.; Duschl, A. Patulin Influences the Expression of Th1/Th2 Cytokines by Activated Peripheral Blood Mononuclear Cells and T Cells Through Depletion of Intracellular Glutathione. Environ. Toxicol. 2008, 23, 84–95. [Google Scholar]
- Marin, M.L.; Murtha, J.; Dong, W.; Pestka, J.J. Effects of mycotoxins on cytokine production and proliferation in EL-4 thymoma cells. J. Toxicol. Environ. Health 1996, 48, 379–396. [Google Scholar]
- Escoula, L.; Thomsen, M.; Bourdiol, D.; Pipy., B.; Peuriere, S.; Roubinet, S. Patulin immunotoxicology: Effect on phagocyte activation and the cellular and humoral immune system of mice and rabbits. Int. J. Immunopharmacol. 1988, 10, 983–989. [Google Scholar]
- Paucod, J.C.; Krivobok, S.; Vidal, D. Immunotoxicity testing of mycotoxins T-2 and patulin on Balb/c mice. Acta Microbiol. Hung. 1990, 37, 331–339. [Google Scholar]
- Escoula, L.; Bourdiol, D.; Linas, M.D.; Recco, P.; Seguela, J.P. Enhancing resistance and modulation of humoral immune response to experimental Candida albicans infection by patulin. Mycopathologia 1988, 103, 153–156. [Google Scholar]
- Llewellyn, G.C.; McCay, J.A.; Brown, R.D.; Musgrove, D.L.; Butterworth, L.F.; Munson, A.E.; White, K.L., Jr. Immunological evaluation of the mycotoxin patulin in female B6C3F1 mice. Food Chem. Toxicol. 1998, 36, 1107–1111. [Google Scholar]
- Mahfoud, R.; Maresca, M.; Garmy, N.; Fantini, J. The mycotoxin patulin alters the barrier function of the intestinal epithelium: Mechanism of action of the toxin and protective effects of glutathione. Toxicol. Appl. Pharmacol. 2002, 181, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Maresca, M.; Yahi, N.; Younès-Sakr, L.; Boyron, M.; Caporiccio, B.; Fantini, J. Both direct and indirect effects account for the pro-inflammatory activity of enteropathogenic mycotoxins on the human intestinal epithelium: Stimulation of interleukin-8 secretion, potentiation of interleukin-1beta effect and increase in the transepithelial passage of commensal bacteria. Toxicol. Appl. Pharmacol. 2008, 228, 84–92. [Google Scholar]
- Bouhet, S.; Oswald, I.P. The effects of mycotoxins, fungal food contaminants, on the intestinal epithelial cell-derived innate immune response. Vet. Immunol. Immunopathol. 2005, 108, 199–209. [Google Scholar]
- Bouhet, S.; Hourcade, E.; Loiseau, N.; Fikry, A.; Martinez, S.; Roselli, M.; Galtier, P.; Mengheri, E.; Oswald, I.P. The mycotoxin fumonisin B1 alters the proliferation and the barrier function of porcine intestinal epithelial cells. Toxicol. Sci. 2004, 77, 165–171. [Google Scholar] [PubMed]
- Pinton, P.; Nougayrède, J.P.; Del Rio, J.C.; Moreno, C.; Marin, D.E.; Ferrier, L.; Bracarense, A.P.; Kolf-Clauw, M.; Oswald, I.P. The food contaminant deoxynivalenol, decreases intestinal barrier permeability and reduces claudin expression. Toxicol. Appl. Pharmacol. 2009, 237, 41–48. [Google Scholar] [Green Version]
- Bayram, O.; Krappmann, S.; Ni, M.; Bok, J.W.; Helmstaedt, K.; Valerius, O.; Braus-Stromeyer, S.; Kwon, N.J.; Keller, N.P.; Yu, J.H.; Braus, G.H. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 2008, 320, 1504–1506. [Google Scholar]
- Duran, R.M.; Cary, J.W.; Calvo, A.M. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 2007, 73, 1158–1168. [Google Scholar]
- Myung, K.; Li, S.; Butchko, R.A.; Busman, M.; Proctor, R.H.; Abbas, H.K.; Calvo, A.M. FvVE1 regulates biosynthesis of the mycotoxins fumonisins and fusarins in Fusarium verticillioides. J. Agric. Food Chem. 2009, 57, 5089–5094. [Google Scholar] [PubMed]
- Bok, J.W.; Keller, N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 2004, 3, 527–535. [Google Scholar]
- Priest, J.W.; Light, R.J. Applications of high-performance liquid chromatography to quantitation of metabolites and enzymes of the patulin pathway from Penicillium patulum. J. Chromatogr. 1990, 513, 237–246. [Google Scholar]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Puel, O.; Galtier, P.; Oswald, I.P. Biosynthesis and Toxicological Effects of Patulin. Toxins 2010, 2, 613-631. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins2040613
Puel O, Galtier P, Oswald IP. Biosynthesis and Toxicological Effects of Patulin. Toxins. 2010; 2(4):613-631. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins2040613
Chicago/Turabian StylePuel, Olivier, Pierre Galtier, and Isabelle P. Oswald. 2010. "Biosynthesis and Toxicological Effects of Patulin" Toxins 2, no. 4: 613-631. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins2040613