A Venom Gland Extracellular Chitin-Binding-Like Protein from Pupal Endoparasitoid Wasps, Pteromalus Puparum, Selectively Binds Chitin
Abstract
:1. Introduction
2. Results
2.1. Full-Length PpCBP cDNA Cloning and Sequence Analysis
| Primer Name | Nucleotide Sequence(5′–3′) |
|---|---|
| PpCBP-5′RACE-outer | ATTAAATTCCGTTCCAGGCCCACAT |
| PpCBP-5′RACE-inner | CGATCAGCCAAGGGCCAGTCACAGA |
| PpCBP-3′RACE-outer | CGATCAGCCAAGGGCCAGTCACAGA |
| PpCBP-3′RACE-inner | ACGAGCACCTTGGCGGTTGCGTTAT |
| PpCBP-SP * | CGCGGATCCGCAACGTACTATCCGTTCCCAG |
| PpCBP-AP * | CCGCTCGAGCTATTTCTTGTTGTAATCGCCATA |
| qPpCBP-SP | ACTATCCGTTCCCAGGTGAC |
| qPpCBP-AP | CATTTTCTGGAAGTCGTC GGA |
| qPp18s-SP | CGAGCGATGAACCGACAG |
| qPp18S-AP | CGGGGAGGTAGTGACGAA |


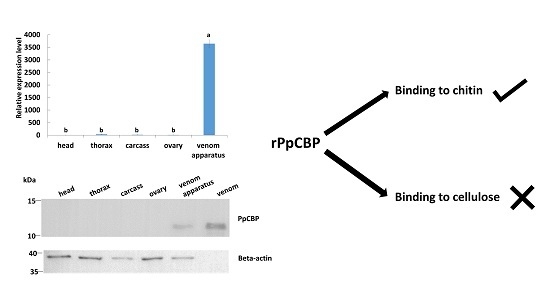
2.2. Tissues and Temporal Expression of PpCBP




2.3. Recombinant PpCBP Expression Products and Cellulose- and Chitin-Binding Capacities


3. Discussion
4. Experimental Section
4.1. Insects and Sample Preparation
4.2. Full-Length cDNAs Cloning
4.3. Sequence Analysis
4.4. Recombinant Expression, Protein Purification, and Antiserum Preparation
4.5. qPCR
4.6. Western Blot
4.7. Immunohistochemistry
4.8. Chitin- and Cellulose-Binding Assay
4.9. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Huang, X.; Xie, W.J.; Gong, Z.Z. Characteristics and antifungal activity of a chitin binding protein from Ginkgo biloba. FEBS Lett. 2000, 478, 123–126. [Google Scholar] [CrossRef]
- Kamakura, T.; Yamaguchi, S.; Saitoh, K.; Teraoka, T.; Yamaguchi, I. A novel gene, CBP1, encoding a putative extracellular chitin-binding protein, may play an important role in the hydrophobic surface sensing of Magnaporthe grisea during appressorium differentiation. Mol. Plant-Microb. Interact. 2002, 15, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Vaaje-Kolstad, G.; Horn, S.J.; van Aalten, D.M.; Synstad, B.; Eijsink, V.G. The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J. Biol. Chem. 2005, 280, 28492–28497. [Google Scholar] [CrossRef] [PubMed]
- Tellam, R.L.; Wijffels, G.; Willadsen, P. Peritrophic matrix proteins. Insect Biochem. Mol. Biol. 1999, 29, 87–101. [Google Scholar] [CrossRef]
- Willis, J.H. Cuticular proteins: The neglected component. Arch. Insect Biochem. Physiol. 1987, 6, 203–215. [Google Scholar] [CrossRef]
- Shen, Z.C.; Jacobs-Lorena, M. Evolution of chitin-binding proteins in invertebrates. J. Mol. Evol. 1999, 48, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Rebers, J.E.; Riddiford, L.M. Structure and expression of a Manduca sexta larval cuticle gene homologous to Drosophila cuticle genes. J. Mol. Biol. 1988, 203, 411–423. [Google Scholar] [CrossRef]
- Willis, J. Cuticular proteins in insects and crustaceans. Integr. Comp. Biol. 1999, 39, 600–609. [Google Scholar] [CrossRef]
- Chae, K.S.; Lee, I.H.; Choi, C.S.; Kim, H.R. Purification and characterization of chitin-binding proteins from the hemolymph of sweet potato hornworm, Agrius convolvuli. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1999, 124, 475–481. [Google Scholar] [CrossRef]
- Asgari, S.; Rivers, D.B. Venom proteins from endoparasitoid wasps and their role in host-parasite interactions. Annu. Rev. Entomol. 2011, 56, 313–335. [Google Scholar] [CrossRef] [PubMed]
- Poirié, M.; Colinet, D.; Gatti, J.L. Insights into function and evolution of parasitoid wasp venoms. Curr. Opin. Insect Sci. 2014, 6, 52–60. [Google Scholar] [CrossRef]
- De Graaf, D.C.; Aerts, M.; Brunain, M.; Desjardins, C.A.; Jacobs, F.J.; Werren, J.H.; Devreese, B. Insights into the venom composition of the ectoparasitoid wasp Nasonia vitripennis from bioinformatic and proteomic studies. Insect Mol. Biol. 2010, 19, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Vincent, B.; Kaeslin, M.; Roth, T.; Heller, M.; Poulain, J.; Cousserans, F.; Schaller, J.; Poirié, M.; Lanzrein, B.; Drezen, J.M. The venom composition of the parasitic wasp Chelonus inanitus resolved by combined expressed sequence tags analysis and proteomic approach. BMC Genom. 2010, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peiren, N.; de Graaf, D.C.; Vanrobaeys, F.; Danneels, E.L.; Devreese, B.; van Beeumen, J.; Jacobs, F.J. Proteomic analysis of the honey bee worker venom gland focusing on the mechanisms of protection against tissue damage. Toxicon 2008, 52, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Ye, G.Y.; Hu, C. Parasitism of Pieris rapae (Lepidoptera: Pieridae) by a pupal endoparasitoid, Pteromalus puparum (Hymenoptera: Pteromalidae): Effects of parasitization and venom on host hemocytes. J. Insect Physiol. 2004, 50, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ye, G.Y.; Cai, J.; Hu, C. Comparative venom toxicity between Pteromalus puparum and Nasonia vitripennis (Hymenoptera: Pteromalidae) toward the hemocytes of their natural hosts, non-target insects and cultured insect cells. Toxicon 2005, 46, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fang, Q.; Qian, C.; Wang, F.; Yu, X.Q.; Ye, G.Y. Inhibition of host cell encapsulation through inhibiting immune gene expression by the parasitic wasp venom calreticulin. Insect Biochem. Mol. Biol. 2013, 43, 936–946. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Wang, L.; Zhu, J.Y.; Li, Y.M.; Song, Q.; Stanley, D.W.; Akhtar, Z.R.; Ye, G.Y. Expression of immune-response genes in lepidopteran host is suppressed by venom from an endoparasitoid, Pteromalus puparum. BMC Genom. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.Y.; Zhu, J.Y.; Zhang, Z.; Fang, Q.; Cai, J.; Hu, C. Venom from the endoparasitoid Pteromalus puparum (Hymenoptera: Pteromalidae) adversely affects host hemocytes: Differential toxicity and microstructural and ultrastructural changes in plasmatocytes and granular cells. In Recent Advances in the Biochemistry, Toxicity, and Mode of Action of Parasitic Wasp Venoms; Rivers, D., Yoder, J., Eds.; Research Singpost: Kerala, India, 2007; pp. 115–127. [Google Scholar]
- Zhu, J.Y.; Ye, G.Y.; Dong, S.Z.; Fang, Q.; Hu, C. Venom of Pteromalus puparum (Hymenoptera: Pteromalidae) induced endocrine changes in the hemolymph of its host, Pieris rapae (Lepidoptera: Pieridae). Arch. Insect Biochem. Physiol. 2009, 71, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.L.; Ye, G.Y.; Zhu, J.Y.; Chen, X.X.; Hu, C. Isolation and characterization of an immunosuppressive protein from venom of the pupa-specific endoparasitoid Pteromalus puparum. J. Invertebr. Pathol. 2008, 99, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Ye, G.Y.; Hu, C. Molecular cloning and characterization of acid phosphatase in venom of the endoparasitoid wasp Pteromalus puparum (Hymenoptera: Pteromalidae). Toxicon 2008, 51, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Ye, G.Y.; Fang, Q.; Hu, C. Alkaline phosphatase from venom of the endoparasitoid wasp, Pteromalus puparum. J. Insect Sci. 2010, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhu, J.Y.; Qian, C.; Fang, Q.; Ye, G.Y. Venom of the parasitoid wasp Pteromalus puparum contains an odorant binding protein. Arch. Insect Biochem. Physiol. 2014, 88, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.C.; Fang, Q.; Wang, L.; Liu, J.D.; Zhu, Y.; Wang, F.; Li, F.; Werren, J.; Ye, G.Y. Insights into the venom composition and evolution of an endoparasitoid wasp by combining proteomic and transcriptomic analyses. Sci. Rep. submitted for publication. 2015. [Google Scholar]
- Fang, Q.; Yan, Z.C.; Werren, J.; Ye, G.Y. Institute of Insect Sciences, Zhejiang University, Hangzhou, China. Unpublished work. 2015. [Google Scholar]
- Rebers, J.E.; Willis, J.H. A conserved domain in arthropod cuticular proteins binds chitin. Insect Biochem. Mol. Biol. 2001, 31, 1083–1093. [Google Scholar] [CrossRef]
- Tetreau, G.; Dittmer, N.T.; Cao, X.L.; Agrawal, S.; Chen, Y.R.; Muthukrishnan, S.; Haobo, J.; Blissard, G.W.; Kanost, M.R.; Wang, P. Analysis of chitin-binding proteins from Manduca sexta provides new insights into evolution of peritrophin A-type chitin-binding domains in insects. Insect Biochem. Mol. Biol. 2014, 62, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Hegedus, D.; Erlandson, M.; Gillott, C.; Gillott, C.; Toprak, U. New insights into peritrophic matrix synthesis, architecture, and function. Annu. Rev. Entomol. 2009, 54, 285–302. [Google Scholar] [CrossRef] [PubMed]
- Devenport, M.; Fujioka, H.; Donnelly-Doman, M.; Shen, Z.; Jacobs-Lorena, M. Storage and secretion of Ag-Aper14, a novel peritrophic matrix protein, and Ag-Muc1 from the mosquito Anopheles gambiae. Cell Tissue Res. 2005, 320, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Wijffels, G.; Eisemann, C.; Riding, G.; Pearson, R.; Jones, A.; Willadsen, P.; Tellam, R. A novel family of chitin-binding proteins from insect type 2 peritrophic matrix cDNA sequences, chitin binding activity, and cellular localization. J. Biol. Chem. 2001, 276, 15527–15536. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Suzuki, M.; Taiyoji, M.; Nikaidou, N.; Watanabe, T. Chitin bonding proetin (CBP21) in the clture supernatant of Serratia marcescens 2170. Biosci. Biotechnol. Biochem. 1998, 62, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Ujita, M.; Sakai, K.; Hamazaki, K.; Yoneda, M.; Isomura, S.; Hara, A. Carbohydrate binding specificity of the recombinant chitin-binding domain of humanmacrophage chitinase. Biosci. Biotechnol. Biochem. 2003, 67, 2402–2407. [Google Scholar] [CrossRef] [PubMed]
- Hardt, M.; Laine, R.A. Mutation of actdive site residues in the chitin-binding domain ChBDChiA1 from chitijnase A1 of Bacillus circulans alters substrate specificity: Use of a green fluorescent protein binding assay. Arch. Biochem. Biophys. 2004, 426, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Jacobs-Lorena, M. Characterization of a novel gut-specific chininase gene from the human malaria vector Anopheles gambiae. J. Biol. Chem. 1998, 273, 17665–17670. [Google Scholar] [CrossRef] [PubMed]
- Van Marle, J.; Piek, T. Morphology of the venom apparatus. In Venoms of the Hymenoptera: Biochemical, Pharmacological and Behavioural Aspects; Piek, T., Ed.; Academic Press: New York, NY, USA, 1986; pp. 17–44. [Google Scholar]
- Andersen, S.O. Biochemistry of insect cuticle. Annu. Rev. Entomol. 1979, 24, 29–59. [Google Scholar] [CrossRef]
- ORF Finder (Open Reading Frame Finder). Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/projects/gorf/ (accessed on 18 August 2015).
- Stanke, M.; Diekhans, M.; Baertsch, R.; Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 2008, 24, 637–644. [Google Scholar] [CrossRef] [PubMed]
- AUGUSTUS Version 2.4. Available online: http://bioinf.uni-greifswald.de/augustus (accessed on 18 August 2015).
- Basic Local Alignment Search Tool (BLAST). Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 18 August 2015).
- InterProScan 5. Available online: http://www.ebi.ac.uk/Tools/pfa/iprscan5/ (accessed on 18 August 2015).
- The signalP 4.0. Available online: http://www.cbs.dtu.dk/services/SignalP-4.0/ (accessed on 18 August 2015).
- ClustalX. Available online: http://www-igbmc.u-strasbg.fr/BioInfo/ClustalX/ (accessed on 18 August 2015).
- MEGA 5.10. Available online: http://www.megasoftware.net/mega.html/ (accessed on 18 August 2015).
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Primer3 Program. Available online: http://primer3.ut.ee/ (accessed on 18 August 2015).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.Y.; Zhang, C.X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2013, 20, 254–260. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Ye, X.-H.; Liu, Y.; Yan, Z.-C.; Stanley, D.; Ye, G.-Y.; Fang, Q. A Venom Gland Extracellular Chitin-Binding-Like Protein from Pupal Endoparasitoid Wasps, Pteromalus Puparum, Selectively Binds Chitin. Toxins 2015, 7, 5098-5113. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins7124867
Zhu Y, Ye X-H, Liu Y, Yan Z-C, Stanley D, Ye G-Y, Fang Q. A Venom Gland Extracellular Chitin-Binding-Like Protein from Pupal Endoparasitoid Wasps, Pteromalus Puparum, Selectively Binds Chitin. Toxins. 2015; 7(12):5098-5113. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins7124867
Chicago/Turabian StyleZhu, Yu, Xin-Hai Ye, Yang Liu, Zhi-Chao Yan, David Stanley, Gong-Yin Ye, and Qi Fang. 2015. "A Venom Gland Extracellular Chitin-Binding-Like Protein from Pupal Endoparasitoid Wasps, Pteromalus Puparum, Selectively Binds Chitin" Toxins 7, no. 12: 5098-5113. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins7124867