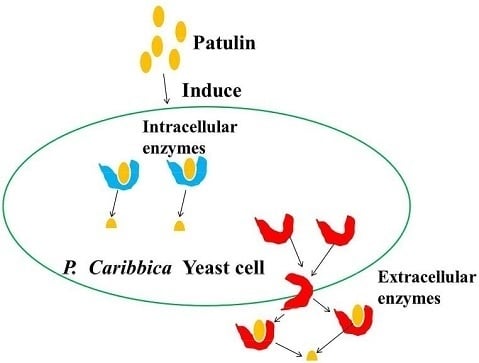
The Possible Mechanisms Involved in Degradation of Patulin by Pichia caribbica
Abstract
:1. Introduction
2. Results
2.1. Effect of Cell-Free Filtrate of P. caribbica on Patulin Degradation
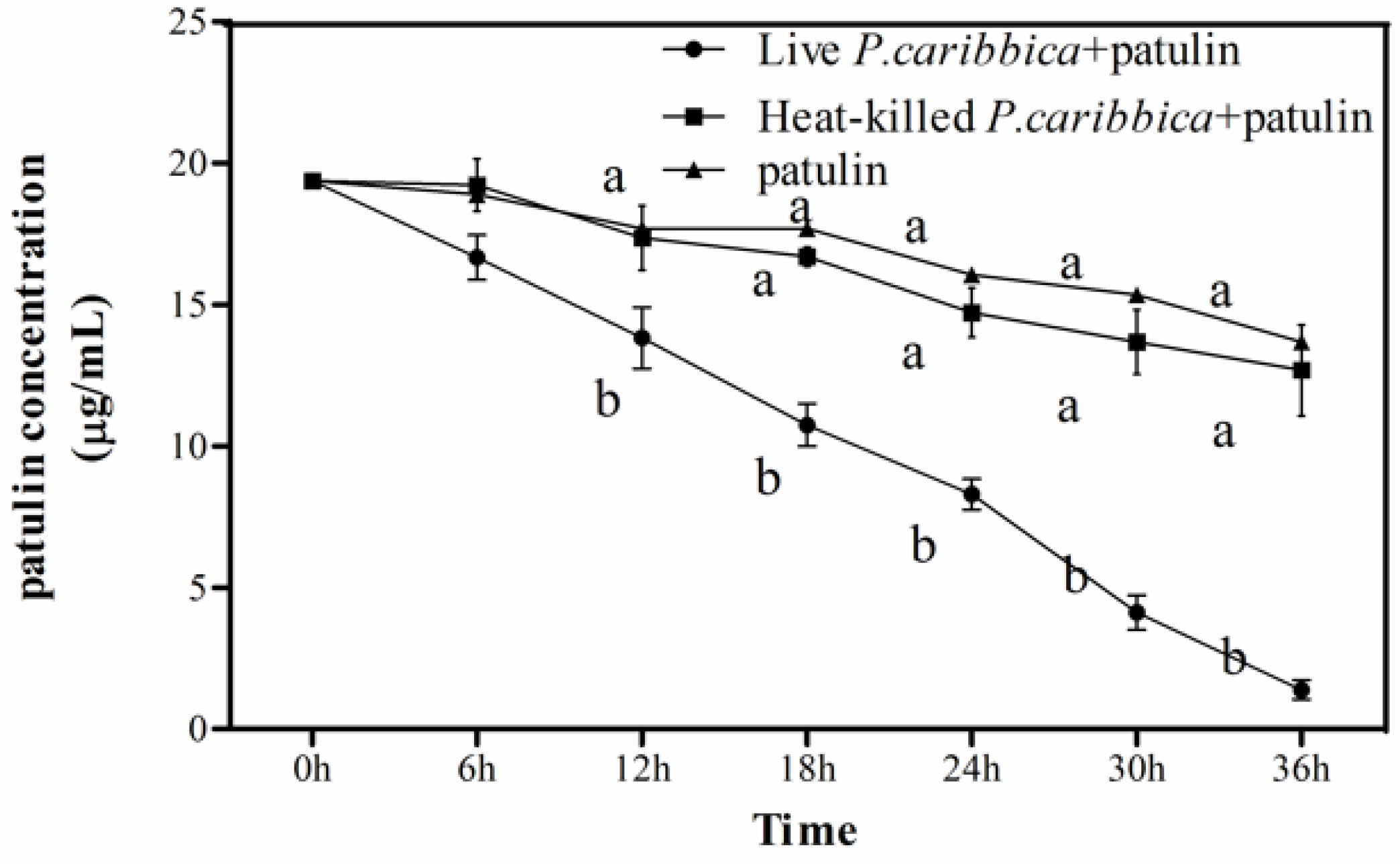
2.2. Effect of Viable and Heat-Killed P. caribbica Cells on Patulin Degradation
2.3. Degradation of Patulin by P. caribbica Was Inhibited by Cycloheximide
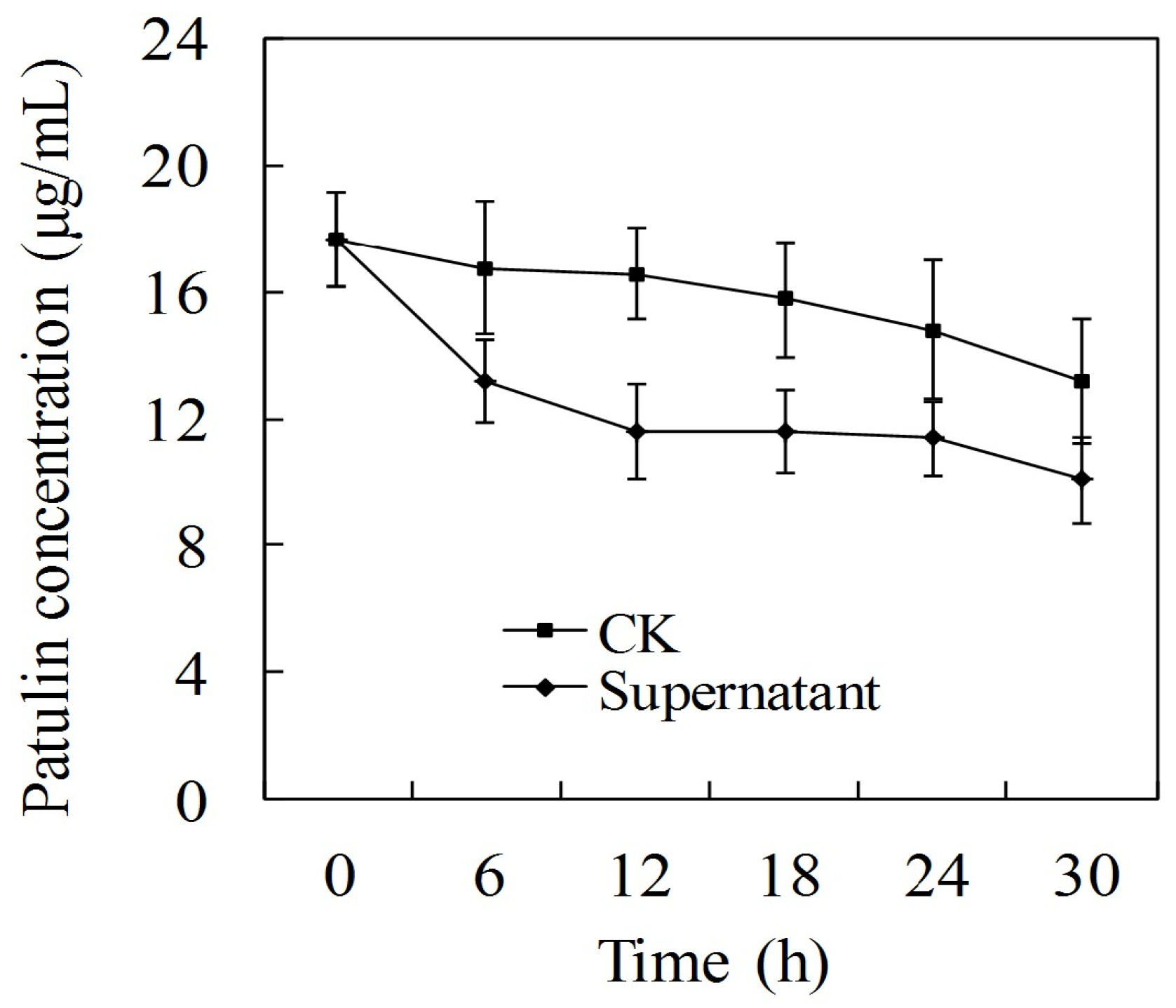
2.4. Effect of P. caribbica’s Supernatant on Patulin Degradation
2.5. Effect of Intracellular Enzymes of P. caribbica on Patulin Degradation
2.6. Identification of Differentially Expressed Proteins
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Antagonist and Growth Conditions
5.2. Preparation of Stock Standard Solutions of Patulin
5.3. HPLC-UV Analysis of Patulin
5.4. Efficacy of Cell-Free Culture Filtrate of P. caribbica on the Detoxification of Patulin
5.5. Efficacy of Viable and Heat-Killed P. caribbica Cells on the Reduction of Patulin
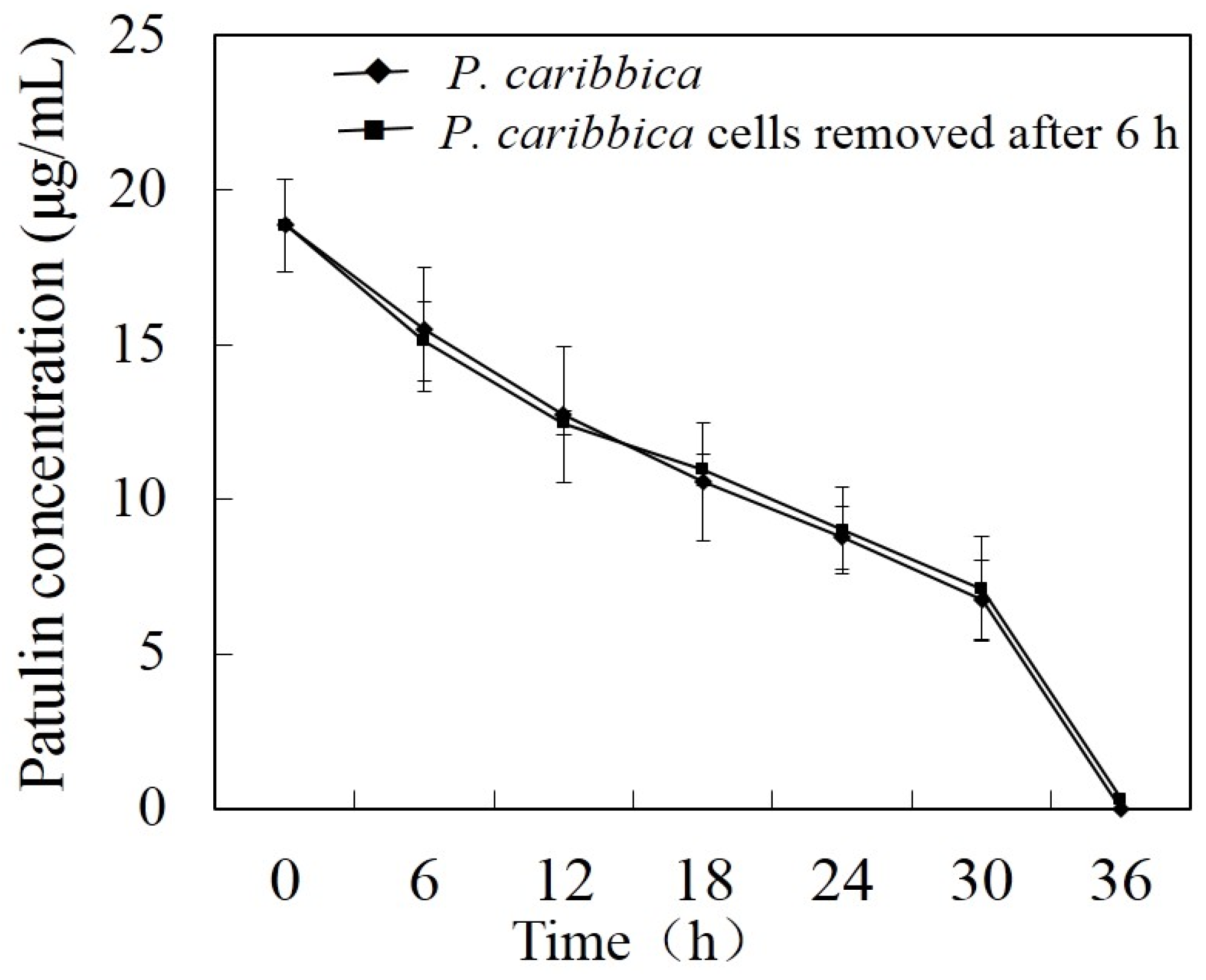
5.6. Absorption of Patulin by P. caribbica Cells
5.7. Effects of Cycloheximide on Degradation of Patulin by P. caribbica
5.8. Effects of P. caribbica Supernatant on Patulin Degradation
5.9. Degradation of Patulin by Intracellular Enzymes
5.10. Protein Sample Preparation
5.11. 2-DE and Image Analysis
5.12. Protein In-Gel Digestion and Identification
5.13. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Joint FAO/WHO Expert Committee on Food Additives; World Health Organization; Food and Agriculture Organization of the United Nations; International Programme on Chemical Safety. Toxicological Evaluation of Certain Food Additives and Contaminants in Food; Cambridge University Press: Cambridge, UK, 1996; pp. 189–197. [Google Scholar]
- Ono, E.Y.S.; Biazon, L.; Silva, M.D.; Vizoni, É.; Sugiura, Y.; Ueno, Y.; Hirooka, E.Y. Fumonisins in corn: Correlation with fusarium sp. Count, damaged kernels, protein and lipid content. Braz. Arch. Biol. Technol. 2006, 49, 1967–1976. [Google Scholar] [CrossRef]
- Pitt, J.I.; Basílico, J.C.; Abarca, M.L.; López, C. Mycotoxins and toxigenic fungi. Med. Mycol. 2010, 38 (Suppl. S1), 41–46. [Google Scholar] [CrossRef]
- Ritieni, A. Patulin in italian commercial apple products. J. Agric. Food Chem. 2003, 51, 6086–6090. [Google Scholar] [CrossRef] [PubMed]
- Andersen, B.; Smedsgaard, J.; Frisvad, J.C. Penicillium expansum: Consistent production of patulin, chaetoglobosins, and other secondary metabolites in culture and their natural occurrence in fruit products. J. Agric. Food Chem. 2004, 52, 2421–2428. [Google Scholar] [CrossRef] [PubMed]
- Waksman, S.A.; Horning, E.S.; Spencer, E.L. The production of two antibacterial substances, fumigacin and clavacin. Am. Assoc. Adv. Sci. 1942, 96, 202–203. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, B. Bactericidal effects of aspergillus clavatus. Nature 1942, 149, 356–357. [Google Scholar] [CrossRef]
- Martins, M.; Gimeno, A.; Martins, H.; Bernardo, F. Co-occurrence of patulin and citrinin in portuguese apples with rotten spots. Food Addit. Contam. 2002, 19, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Barreira, M.J.; Alvito, P.C.; Almeida, C.M. Occurrence of patulin in apple-based-foods in portugal. Food Chem. 2010, 121, 653–658. [Google Scholar] [CrossRef]
- Baert, K.; de Meulenaer, B.; Kamala, A.; Kasase, C.; Devlieghere, F. Occurrence of patulin in organic, conventional, and handcrafted apple juices marketed in belgium. J. Food Prot. 2006, 69, 1371–1378. [Google Scholar] [PubMed]
- Saxena, N.; Dwivedi, P.D.; Ansari, K.M.; Das, M. Patulin in apple juices: Incidence and likely intake in an indian population. Food Addit. Contam. 2008, 1, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhuang, H.; Zhang, T.; Liu, J. Patulin content in apple products marketed in northeast china. Food Control 2010, 21, 1488–1491. [Google Scholar] [CrossRef]
- Schumacher, D.M.; Metzler, M.; Lehmann, L. Mutagenicity of the mycotoxin patulin in cultured chinese hamster v79 cells, and its modulation by intracellular glutathione. Arch. Toxicol. 2005, 79, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Osswald, H.; Frank, H.; Komitowski, D.; Winter, H. Long-term testing of patulin administered orally to sprague-dawley rats and swiss mice. Food Cosmet. Toxicol. 1976, 16, 243–247. [Google Scholar] [CrossRef]
- Wichmann, G.; Herbarth, O.; Lehmann, I. The mycotoxins citrinin, gliotoxin, and patulin affect interferon-γ rather than interleukin-4 production in human blood cells. Environ. Toxicol. 2002, 17, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, H.; Radha, S.K.; Shanmugasundaram, E. Neurotoxic effect of patulin. Indian J. Exp. Biol. 1982, 20, 230–231. [Google Scholar] [PubMed]
- Ciegler, A.; Beckwith, A.; Jackson, L.K. Teratogenicity of patulin and patulin adducts formed with cysteine. Appl. Environ. Microbiol. 1976, 31, 664–667. [Google Scholar] [PubMed]
- Donmez-Altuntas, H.; Gokalp-Yildiz, P.; Bitgen, N.; Hamurcu, Z. Evaluation of genotoxicity, cytotoxicity and cytostasis in human lymphocytes exposed to patulin by using the cytokinesis-block micronucleus cytome (CBMN cyt) assay. Mycotoxin Res. 2013, 29, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Nunes, C.A. Biological control of postharvest diseases of fruit. Eur. J. Plant Pathol. 2012, 133, 181–196. [Google Scholar] [CrossRef]
- Droby, S.; Wisniewski, M.; Macarisin, D.; Wilson, C. Twenty years of postharvest biocontrol research: Is it time for a new paradigm? Postharvest Biol. Technol. 2009, 52, 137–145. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, H.; Yang, Q.; Ren, R. Efficacy of pichia caribbica in controlling blue mold rot and patulin degradation in apples. Int. J. Food Microbiol. 2013, 162, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Castoria, R.; Morena, V.; Caputo, L.; Panfili, G.; de Curtis, F.; De Cicco, V. Effect of the biocontrol yeast rhodotorula glutinis strain LS11 on patulin accumulation in stored apples. Phytopathology 2005, 95, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Coelho, A.; Celli, M.; Sataque Ono, E.; Hoffmann, F.; Pagnocca, F.; Garcia, S.; Sabino, M.; Harada, K.; Wosiacki, G.; Hirooka, E. Patulin biodegradation using Pichia ohmeri and Saccharomyces cerevisiae. World Mycotoxin J. 2008, 1, 325–331. [Google Scholar] [CrossRef]
- Castoria, R.; Mannina, L.; Durán-Patrón, R.; Maffei, F.; Sobolev, A.P.; de Felice, D.V.; Pinedo-Rivilla, C.; Ritieni, A.; Ferracane, R.; Wright, S.A. Conversion of the mycotoxin patulin to the less toxic desoxypatulinic acid by the biocontrol yeast Rhodosporidium kratochvilovae strain LS11. J. Agric. Food Chem. 2011, 59, 11571–11578. [Google Scholar] [CrossRef] [PubMed]
- Bevardi, M.; Frece, J.; Mesarek, D.; Bošnir, J.; Mrvčić, J.; Delaš, F.; Markov, K. Antifungal and antipatulin activity of Gluconobacter oxydans isolated from apple surface. Arch. Ind. Hyg. Toxicol. 2013, 64, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Coelho, A.R.; Celli, M.G.; Ono, E.Y.S.; Wosiacki, G.; Hoffmann, F.L.; Pagnocca, F.C.; Hirooka, E.Y. Penicillium expansum versus antagonist yeasts and patulin degradation in vitro. Braz. Arch. Biol. Technol. 2007, 50, 725–733. [Google Scholar] [CrossRef]
- Zhu, R.; Yu, T.; Guo, S.; Hu, H.; Zheng, X.; Karlovsky, P. Effect of the yeast Rhodosporidium paludigenum on postharvest decay and patulin accumulation in apples and pears. J. Food Prot. 2015, 78, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Feussner, K.; Wu, T.; Yan, F.; Karlovsky, P.; Zheng, X. Detoxification of mycotoxin patulin by the yeast Rhodosporidium paludigenum. Food Chem. 2015, 179, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Iwahashi, Y.; Hosoda, H.; Park, J.-H.; Lee, J.-H.; Suzuki, Y.; Kitagawa, E.; Murata, S.M.; Jwa, N.-S.; Gu, M.-B.; Iwahashi, H. Mechanisms of patulin toxicity under conditions that inhibit yeast growth. J. Agric. Food Chem. 2006, 54, 1936–1942. [Google Scholar] [CrossRef] [PubMed]
- Ianiri, G.; Idnurm, A.; Wright, S.A.; Durán-Patrón, R.; Mannina, L.; Ferracane, R.; Ritieni, A.; Castoria, R. Searching for genes responsible for patulin degradation in a biocontrol yeast provides insight into the basis for resistance to this mycotoxin. Appl. Environ. Microbiol. 2013, 79, 3101–3115. [Google Scholar] [CrossRef] [PubMed]
- Ianiri, G.; Idnurm, A.; Castoria, R. Transcriptomic responses of the basidiomycete yeast sporobolomyces sp. To the mycotoxin patulin. BMC Genom. 2016, 17, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Di, S.V.; Pitonzo, R.; Cicero, N.; D’Oca, M.C. Mycotoxin contamination of animal feedingstuff: Detoxification by gamma-irradiation and reduction of aflatoxins and ochratoxin A concentrations. Food Addit. Contam. 2014, 31, 2034–2039. [Google Scholar]
- Shi, L.; Liang, Z.; Li, J.; Hao, J.; Xu, Y.; Huang, K.; Tian, J.; He, X.; Xu, W. Ochratoxin A biocontrol and biodegradation by Bacillus subtilis cw 14. J. Sci. Food Agric. 2014, 94, 1879–1885. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Jiang, W.; Li, C.; Ma, N.; Xu, Y.; Meng, X. Patulin biodegradation by marine yeast kodameae ohmeri. Food Additives & Contaminants Part A Chemistry Analysis Control Exposure & Risk Assessment 2015, 32, 352–360. [Google Scholar]
- Zhang, H.; Dong, M.; Yang, Q.; Tibiru, A.M.; Li, J.; Zhang, X. Biodegradation of zearalenone by Saccharomyces cerevisiae: Possible involvement of zen responsive proteins of the yeast. J. Proteomics 2016, 143, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Folger, B.C. Patulin degradation by yeast protein extract. Master’s Theses, University of Minnesota, Minneapolis, MN, 2014. [Google Scholar]
- Majoul, T.; Bancel, E.; Triboï, E.; Ben Hamida, J.; Branlard, G. Proteomic analysis of the effect of heat stress on hexaploid wheat grain: Characterization of heat-responsive proteins from total endosperm. Proteomics 2003, 3, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, S.K.; Murshid, A.; Prince, T. The shock of aging: Molecular chaperones and the heat shock response in longevity and aging–a mini-review. Gerontology 2009, 55, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, R.I. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008, 22, 1427–1438. [Google Scholar] [CrossRef] [PubMed]
- Voellmy, R.; Boellmann, F. Chaperone regulation of the heat shock protein response. Adv. Exp. Med. Biol. 2007, 23, 89–99. [Google Scholar]
- Bukau, B.; Deuerling, E.; Pfund, C.; Craig, E.A. Getting newly synthesized proteins into shape. Cell 2000, 101, 119–122. [Google Scholar] [CrossRef]
- Hartl, F.U.; Hayer-Hartl, M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 2002, 295, 1852–1858. [Google Scholar] [CrossRef] [PubMed]
- Young, J.C.; Barral, J.M.; Hartl, F.U. More than folding: Localized functions of cytosolic chaperones. Trends Biochem. Sci. 2003, 28, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Pratt, W.B.; Toft, D.O. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 2003, 228, 111–133. [Google Scholar]
- Kepes, F.; Schekman, R. The yeast sec53 gene encodes phosphomannomutase. J. Biol. Chem. 1988, 263, 9155–9161. [Google Scholar] [PubMed]
- Seifert, G.J. Nucleotide sugar interconversions and cell wall biosynthesis: How to bring the inside to the outside. Curr. Opin. Plant Biol. 2004, 7, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Lerouge, P.; Cabanes-Macheteau, M.; Rayon, C.; Fischette-Lainé, A.-C.; Gomord, V.; Faye, L. N-glycoprotein biosynthesis in plants: Recent developments and future trends. Plant Mol. Biol. 1998, 38, 31–48. [Google Scholar] [CrossRef] [PubMed]
- Spiro, R.G. Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 2002, 12, 43R–56R. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.; Fell, J.; Boekhout, T. Definition, classification and nomenclature of the yeasts. The yeasts, a taxonomic study 2011, 1, 3–5. [Google Scholar]
- Li, S.-S.; Cheng, C.; Li, Z.; Chen, J.-Y.; Yan, B.; Han, B.-Z.; Reeves, M. Yeast species associated with wine grapes in china. Int. J. Food Microbiol. 2010, 138, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.F. P. caribbica shown to be safe in physiological, acute toxicity and Ames test in animal. Jiangsu University: Zhenjiang, China, Unpublished work. 2016. [Google Scholar]
- MacDonald, S.; Long, M.; Gilbert, J.; Felgueiras, I. Liquid chromatographic method for determination of patulin in clear and cloudy apple juices and apple puree: Collaborative study. J. AOAC Int. 2000, 83, 1387–1394. [Google Scholar] [PubMed]
- Li, B.; Lai, T.; Qin, G.; Tian, S. Ambient ph stress inhibits spore germination of penicillium expansum by impairing protein synthesis and folding: A proteomic-based study. J. Proteome Res. 2010, 9, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, L.H.; Li, Q.; Ma, Z.; Chu, C. Differential proteomic analysis of proteins in wheat spikes induced by fusarium graminearum. Proteomics 2005, 5, 4496–4503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, Z.; Jiang, L.; Jiang, J.; Luo, H.; Fu, L. Effect of post-harvest heat treatment on proteome change of peach fruit during ripening. J. Proteom. 2011, 74, 1135–1149. [Google Scholar] [CrossRef] [PubMed]


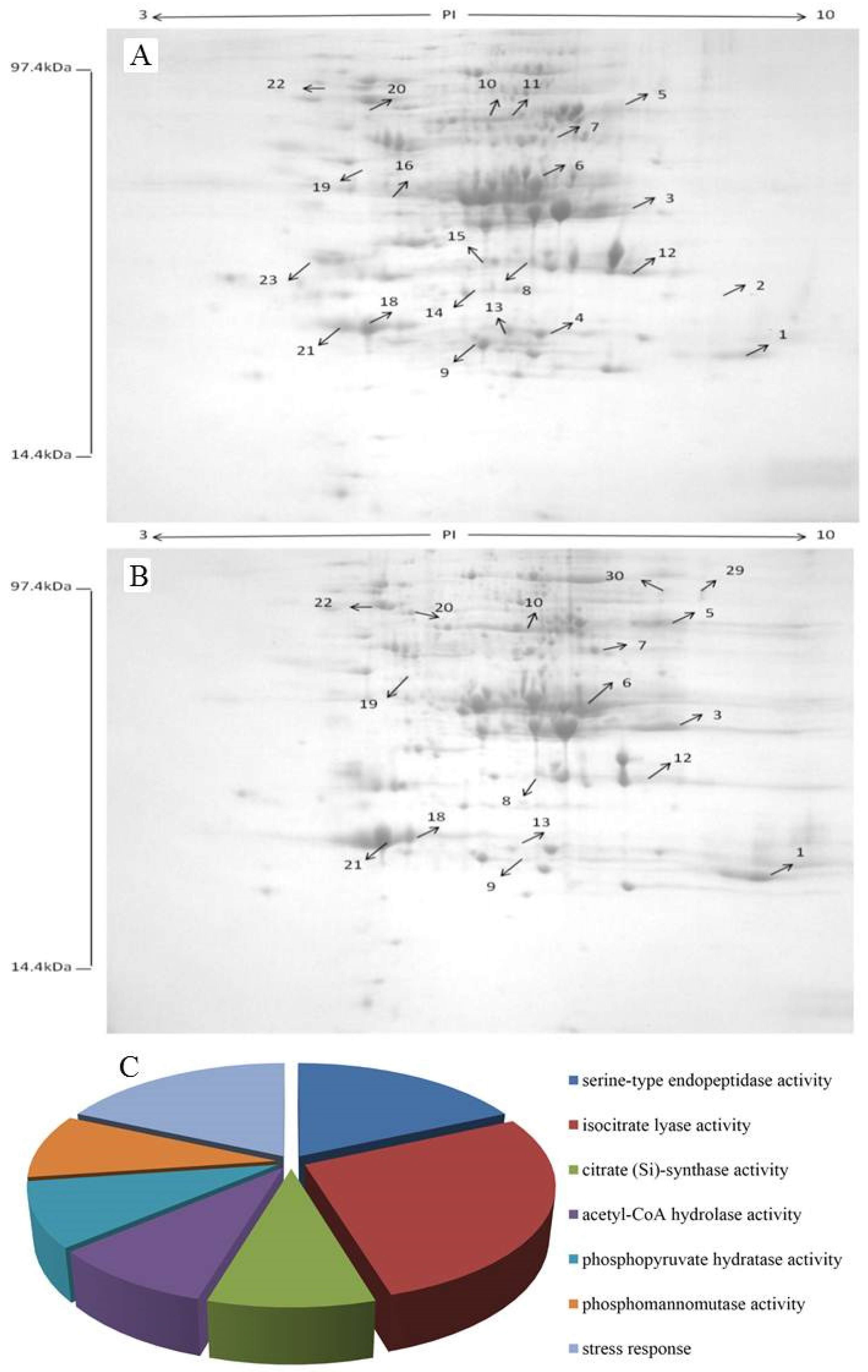
| Molecular Function | Spot | Protein Name | NCBI Accession | Mass | PI | Species | Score | Sequence Coverage (%) | Number of Mass Values Matched |
|---|---|---|---|---|---|---|---|---|---|
| Serine-type endopeptidase activity | 2 | Chymotrypsinogen A | gi|117615 | 26,220 | 8.52 | Bos taurus | 206 | 16 | 4 |
| Serine-type endopeptidase activity | 21 | Chymotrypsinogen | gi|117616 | 26,309 | 4.99 | Bos taurus | 110 | 6 | 1 |
| Isocitrate lyase activity | 4 | Isocitrate lyase | gi|146413757 | 61,937 | 6.31 | Meyerozyma guilliermondii ATCC 6260 | 124 | 5 | 2 |
| Isocitrate lyase activity | 8 | Isocitrate lyase | gi|146413757 | 61,937 | 6.31 | Meyerozyma guilliermondii ATCC 6260 | 429 | 12 | 5 |
| Isocitrate lyase activity | 11 | Isocitrate lyase and phosphorylmutase | gi|344232420 | 62,282 | 6.78 | Candida tenuis ATCC 10573 | 67 | 5 | 2 |
| Citratesynthase activity | 6 | Citrate synthase | gi|146421975 | 43,995 | 6.25 | Meyerozyma guilliermondii ATCC 6260 | 119 | 5 | 2 |
| Acetyl-CoA hydrolase activity | 10 | Acetyl-CoA hydrolase | gi|146417797 | 58,385 | 5.96 | Meyerozyma guilliermondii ATCC 6260 | 222 | 6 | 2 |
| Phosphopyruvate hydratase activity | 15 | Enolase 1 | gi|146415384 | 46,951 | 5.42 | Meyerozyma guilliermondii ATCC 6260 | 117 | 3 | 1 |
| Phosphomannomutase activity | 17 | Phosphomannomutase | gi|146423739 | 28,678 | 5.26 | Meyerozyma guilliermondii ATCC 6260 | 361 | 19 | 5 |
| Stress response | 20 | Heat shock protein SSB1 | gi|146420661 | 66,421 | 5.29 | Meyerozyma guilliermondii ATCC 6260 | 260 | 10 | 5 |
| Stress response | 22 | Heat shock protein 70 2 | gi|146413777 | 70,177 | 5.04 | Meyerozyma guilliermondii ATCC 6260 | 951 | 17 | 12 |
| Unclassified | 12 | DEHA2F04796p | gi|50423973 | 35,926 | 6.24 | Debaryomyces hansenii CBS767 | 61 | 7 | 2 |
| Unclassified | 13 | Conserved hypothetical protein | gi|146417765 | 34,916 | 7.17 | Meyerozyma guilliermondii ATCC 6260 | 219 | 13 | 3 |
| Unclassified | 14 | Hypothetical protein PGUG_05640 | gi|146413298 | 65,256 | 5.85 | Meyerozyma guilliermondii ATCC 6260 | 170 | 5 | 2 |
| Unclassified | 16 | DEHA2G14058p | gi|50427089 | 47,210 | 5.28 | Debaryomyces hansenii CBS767 | 197 | 7 | 2 |
| Unclassified | 5 | Hypothetical protein PGUG_05024 | gi|146414197 | 32,118 | 7.77 | Meyerozyma guilliermondii ATCC 6260 | 122 | 10 | 2 |
| Unclassified | 9 | DEHA2D06160p | gi|50420381 | 54,282 | 5.68 | Debaryomyces hansenii CBS767 | 133 | 8 | 2 |
| Unclassified | 23 | Conserved hypothetical protein | gi|146416825 | 44,146 | 5.33 | Meyerozyma guilliermondii ATCC 6260 | 280 | 13 | 4 |
| Unclassified | 24 | Hypothetical protein PGUG_00755 | gi|146422888 | 20,317 | 5.45 | Meyerozyma guilliermondii ATCC 6260 | 84 | 6 | 1 |
| Unclassified | 25 | Hypothetical protein PGUG_04067 | gi|146414736 | 34,234 | 6.64 | Meyerozyma guilliermondii ATCC 6260 | 104 | 11 | 3 |
| Unclassified | 24 | Hypothetical protein PGUG_03175 | gi|146418962 | 62,015 | 5.21 | Meyerozyma guilliermondii ATCC 6260 | 272 | 12 | 4 |
| Unclassified | 29 | Hypothetical protein PGUG_00646 | gi|190344794 | 78,744 | 8.19 | Meyerozyma guilliermondii ATCC 6260 | 296 | 11 | 6 |
| Unclassified | 30 | Hypothetical protein LOC100448380 | gi|297701166 | 105,979 | 5.19 | Pongo abelii | 85 | 3 | 3 |
| Unclassified | 31 | Hypothetical protein PGUG_03973 | gi|146414548 | 38,139 | 5.53 | Meyerozyma guilliermondii ATCC 6260 | 127 | 5 | 1 |
| Unclassified | 32 | Hypothetical protein PGUG_01788 | gi|146420321 | 37,067 | 5.56 | Meyerozyma guilliermondii ATCC 6260 | 109 | 9 | 2 |
| Unclassified | 33 | Hypothetical protein PGUG_00294 | gi|146421948 | 35,991 | 5.22 | Meyerozyma guilliermondii ATCC 6260 | 88 | 8 | 2 |
| Unclassified | 1 | Chain Z | gi|230350 | 24,662 | 8.23 | Bos taurus | 316 | 29 | 6 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, X.; Yang, Q.; Zhang, H.; Cao, J.; Zhang, X.; Apaliya, M.T. The Possible Mechanisms Involved in Degradation of Patulin by Pichia caribbica. Toxins 2016, 8, 289. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins8100289
Zheng X, Yang Q, Zhang H, Cao J, Zhang X, Apaliya MT. The Possible Mechanisms Involved in Degradation of Patulin by Pichia caribbica. Toxins. 2016; 8(10):289. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins8100289
Chicago/Turabian StyleZheng, Xiangfeng, Qiya Yang, Hongyin Zhang, Jing Cao, Xiaoyun Zhang, and Maurice Tibiru Apaliya. 2016. "The Possible Mechanisms Involved in Degradation of Patulin by Pichia caribbica" Toxins 8, no. 10: 289. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins8100289