Perfringolysin O Theta Toxin as a Tool to Monitor the Distribution and Inhomogeneity of Cholesterol in Cellular Membranes
Abstract
:1. Introduction
2. Structure and Mutations of PFO
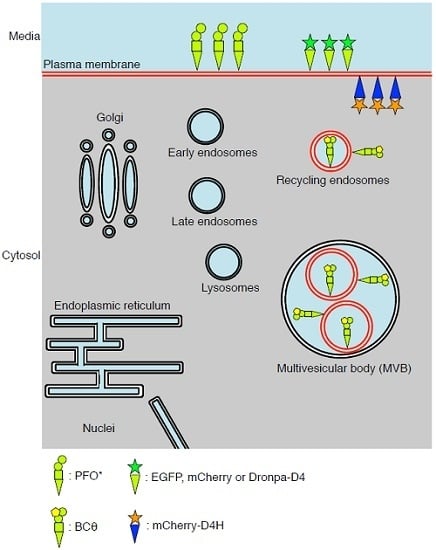
3. PFO*, a Non-Toxic PFO, for Detection of Cholesterol in the Exofacial Leaflets of the Plasma Membrane
4. BCθ, a Biotinylated PFO Fragment for Visualization of Cholesterol in the Endocytic Pathway
5. D4 for the Visualization of Cholesterol Domains in the Exofacial Leaflets of the Plasma Membrane
6. D4H for Visualization of Cholesterol in the Cytosolic Leaflets of the Cellular Membranes
7. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tweten, R.K.; Parker, M.W.; Johnson, A.E. The cholesterol-dependent cytolysins. Curr. Top. Microbiol. Immunol. 2001, 257, 15–33. [Google Scholar] [PubMed]
- Tweten, R.K. Cholesterol-dependent cytolysins, a family of versatile pore-forming toxins. Infect. Immun. 2005, 73, 6199–6209. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, R.J. Cholesterol-dependent cytolysins. Adv. Exp. Med. Biol. 2010, 677, 56–66. [Google Scholar] [PubMed]
- Giddings, K.S.; Zhao, J.; Sims, P.J.; Tweten, R.K. Human CD59 is a receptor for the cholesterol-dependent cytolysin intermedilysin. Nat. Struct. Mol. Biol. 2004, 11, 1173–1178. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.B.; Heuck, A.P. Perfringolysin O structure and mechanism of pore formation as a paradigm for cholesterol-dependent cytolysins. Subcellular Biochem. 2014, 80, 63–81. [Google Scholar]
- Olofsson, A.; Hebert, H.; Thelestam, M. The projection structure of Perfringolysin O (Clostridium perfringens θ-toxin). FEBS Lett. 1993, 319, 125–127. [Google Scholar] [CrossRef]
- Morgan, P.J.; Hyman, S.C.; Rowe, A.J.; Mitchell, T.J.; Andrew, P.W.; Saibil, H.R. Subunit organisation and symmetry of pore-forming, oligomeric pneumolysin. FEBS Lett. 1995, 371, 77–80. [Google Scholar] [CrossRef]
- Heuck, A.P.; Tweten, R.K.; Johnson, A.E. β-barrel pore-forming toxins: Intriguing dimorphic proteins. Biochemistry 2001, 40, 9065–9073. [Google Scholar] [CrossRef] [PubMed]
- Shatursky, O.; Heuck, A.P.; Shepard, L.A.; Rossjohn, J.; Parker, M.W.; Johnson, A.E.; Tweten, R.K. The mechanism of membrane insertion for a cholesterol-dependent cytolysin: A novel paradigm for pore-forming toxins. Cell 1999, 99, 293–299. [Google Scholar] [CrossRef]
- Dang, T.X.; Milligan, R.A.; Tweten, R.K.; Wilson-Kubalek, E.M. Helical crystallization on nickel-lipid nanotubes: Perfringolysin O as a model protein. J. Struct. Biol. 2005, 152, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, R.; Heuck, A.P.; Tweten, R.K.; Johnson, A.E. Structural insights into the membrane-anchoring mechanism of a cholesterol-dependent cytolysin. Nat. Struct. Biol. 2002, 9, 823–827. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.K.; Tweten, R.K.; Johnson, A.E. Disulfide-bond scanning reveals assembly state and β-strand tilt angle of the PFO β-barrel. Nat. Chem. Biol. 2013, 9, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Tweten, R.K. Nucleotide sequence of the gene for Perfringolysin O (θ-toxin) from Clostridium perfringens: Significant homology with the genes for streptolysin O and pneumolysin. Infect. Immun. 1988, 56, 3235–3240. [Google Scholar] [PubMed]
- Ohno-Iwashita, Y.; Iwamoto, M.; Mitsui, K.; Ando, S.; Nagai, Y. Protease-nicked θ-toxin of Clostridium perfringens, a new membrane probe with no cytolytic effect, reveals two classes of cholesterol as toxin-binding sites on sheep erythrocytes. Eur. J. Biochem. 1988, 176, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Ohno-Iwashita, Y.; Iwamoto, M.; Ando, S.; Iwashita, S. Effect of lipidic factors on membrane cholesterol topology—Mode of binding of θ-toxin to cholesterol in liposomes. Biochim. Biophys. Acta 1992, 1109, 81–90. [Google Scholar] [CrossRef]
- Rossjohn, J.; Feil, S.C.; McKinstry, W.J.; Tweten, R.K.; Parker, M.W. Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form. Cell 1997, 89, 685–692. [Google Scholar] [CrossRef]
- Hotze, E.M.; Wilson-Kubalek, E.M.; Rossjohn, J.; Parker, M.W.; Johnson, A.E.; Tweten, R.K. Arresting pore formation of a cholesterol-dependent cytolysin by disulfide trapping synchronizes the insertion of the transmembrane β-sheet from a prepore intermediate. J. Biol. Chem. 2001, 276, 8261–8268. [Google Scholar] [CrossRef] [PubMed]
- Heuck, A.P.; Savva, C.G.; Holzenburg, A.; Johnson, A.E. Conformational changes that effect oligomerization and initiate pore formation are triggered throughout Perfringolysin O upon binding to cholesterol. J. Biol. Chem. 2007, 282, 22629–22637. [Google Scholar] [CrossRef] [PubMed]
- Nelson, L.D.; Johnson, A.E.; London, E. How interaction of Perfringolysin O with membranes is controlled by sterol structure, lipid structure, and physiological low pH: Insights into the origin of Perfringolysin O-lipid raft interaction. J. Biol. Chem. 2008, 283, 4632–4642. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.B.; Moe, P.C.; Wang, D.; Rossi, K.; Trigatti, B.L.; Heuck, A.P. Modifications in Perfringolysin O domain 4 alter the cholesterol concentration threshold required for binding. Biochemistry 2012, 51, 3373–3382. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, A.; Sekizawa, Y.; Emoto, K.; Sakuraba, H.; Inoue, K.; Kobayashi, H.; Umeda, M. Lysenin, a novel sphingomyelin-specific binding protein. J. Biol. Chem. 1998, 273, 5300–5306. [Google Scholar] [CrossRef] [PubMed]
- Bakrac, B.; Kladnik, A.; Macek, P.; McHaffie, G.; Werner, A.; Lakey, J.H.; Anderluh, G. A toxin-based probe reveals cytoplasmic exposure of Golgi sphingomyelin. J. Biol. Chem. 2010, 285, 22186–22195. [Google Scholar] [CrossRef] [PubMed]
- Yachi, R.; Uchida, Y.; Balakrishna, B.H.; Anderluh, G.; Kobayashi, T.; Taguchi, T.; Arai, H. Subcellular localization of sphingomyelin revealed by two toxin-based probes in mammalian cells. Genes Cells 2012, 17, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Makino, A.; Abe, M.; Murate, M.; Inaba, T.; Yilmaz, N.; Hullin-Matsuda, F.; Kishimoto, T.; Schieber, N.L.; Taguchi, T.; Arai, H.; et al. Visualization of the heterogeneous membrane distribution of sphingomyelin associated with cytokinesis, cell polarity, and sphingolipidosis. FASEB J. 2015, 29, 477–493. [Google Scholar] [CrossRef] [PubMed]
- Sandvig, K.; Olsnes, S.; Brown, J.E.; Petersen, O.W.; van Deurs, B. Endocytosis from coated pits of Shiga toxin: A glycolipid-binding protein from Shigella dysenteriae 1. J. Cell. Biol. 1989, 108, 1331–1343. [Google Scholar] [CrossRef] [PubMed]
- Orlandi, P.A.; Fishman, P.H. Orientation of cholera toxin bound to target cells. J. Biol. Chem. 1993, 268, 17038–17044. [Google Scholar] [PubMed]
- Singer, S.J.; Nicolson, G.L. The fluid mosaic model of the structure of cell membranes. Science 1972, 175, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Ikonen, E. Cellular cholesterol trafficking and compartmentalization. Nat. Rev. Mol. Cell Biol. 2008, 9, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, A.; McConnell, H.M. Condensed complexes of cholesterol and phospholipids. Biophys. J. 1999, 77, 1507–1517. [Google Scholar] [CrossRef]
- McConnell, H.M.; Radhakrishnan, A. Condensed complexes of cholesterol and phospholipids. Biochim. Biophys. Acta 2003, 1610, 159–173. [Google Scholar] [CrossRef]
- Ohvo-Rekila, H.; Ramstedt, B.; Leppimaki, P.; Slotte, J.P. Cholesterol interactions with phospholipids in membranes. Prog. Lipid Res. 2002, 41, 66–97. [Google Scholar] [CrossRef]
- Lingwood, D.; Simons, K. Lipid rafts as a membrane-organizing principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Toomre, D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000, 1, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Ehehalt, R. Cholesterol, lipid rafts, and disease. J. Clin. Invest. 2002, 110, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Chadda, R.; Howes, M.T.; Plowman, S.J.; Hancock, J.F.; Parton, R.G.; Mayor, S. Cholesterol-sensitive Cdc42 activation regulates actin polymerization for endocytosis via the GEEC pathway. Traffic 2007, 8, 702–717. [Google Scholar] [CrossRef] [PubMed]
- Mayor, S.; Sabharanjak, S.; Maxfield, F.R. Cholesterol-dependent retention of GPI-anchored proteins in endosomes. EMBO J. 1998, 17, 4626–4638. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, A.; Sharma, D.K.; Marks, D.L.; Pagano, R.E. Elevated endosomal cholesterol levels in niemann-pick cells inhibit Rab4 and perturb membrane recycling. Mol. Biol. Cell 2004, 15, 4500–4511. [Google Scholar] [CrossRef] [PubMed]
- Balse, E.; El-Haou, S.; Dillanian, G.; Dauphin, A.; Eldstrom, J.; Fedida, D.; Coulombe, A.; Hatem, S.N. Cholesterol modulates the recruitment of Kv1.5 channels from Rab11-associated recycling endosome in native atrial myocytes. Proc. Natl. Acad Sci. USA 2009, 106, 14681–14686. [Google Scholar] [CrossRef] [PubMed]
- Kozik, P.; Hodson, N.A.; Sahlender, D.A.; Simecek, N.; Soromani, C.; Wu, J.; Collinson, L.M.; Robinson, M.S. A human genome-wide screen for regulators of clathrin-coated vesicle formation reveals an unexpected role for the V-ATPase. Nat. Cell Biol. 2013, 15, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Reverter, M.; Rentero, C.; Garcia-Melero, A.; Hoque, M.; Vila de Muga, S.; Alvarez-Guaita, A.; Conway, J.R.; Wood, P.; Cairns, R.; Lykopoulou, L.; et al. Cholesterol regulates syntaxin 6 trafficking at trans-Golgi network endosomal boundaries. Cell Rep. 2014, 7, 883–897. [Google Scholar] [CrossRef] [PubMed]
- Gagescu, R.; Demaurex, N.; Parton, R.G.; Hunziker, W.; Huber, L.A.; Gruenberg, J. The recycling endosome of madin-darby canine kidney cells is a mildly acidic compartment rich in raft components. Mol. Biol. Cell 2000, 11, 2775–2791. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, M.; Fairn, G.D. Complementary probes reveal that phosphatidylserine is required for the proper transbilayer distribution of cholesterol. J. Cell Sci. 2015, 128, 1422–1433. [Google Scholar] [CrossRef] [PubMed]
- Holtta-Vuori, M.; Tanhuanpaa, K.; Mobius, W.; Somerharju, P.; Ikonen, E. Modulation of cellular cholesterol transport and homeostasis by Rab11. Mol. Biol. Cell 2002, 13, 3107–3122. [Google Scholar] [CrossRef] [PubMed]
- Alpy, F.; Tomasetto, C. Give lipids a start: The star-related lipid transfer (start) domain in mammals. J. Cell Sci. 2005, 118, 2791–2801. [Google Scholar] [CrossRef] [PubMed]
- Mesmin, B.; Pipalia, N.H.; Lund, F.W.; Ramlall, T.F.; Sokolov, A.; Eliezer, D.; Maxfield, F.R. Stard4 abundance regulates sterol transport and sensing. Mol. Biol. Cell 2011, 22, 4004–4015. [Google Scholar] [CrossRef] [PubMed]
- Garbarino, J.; Pan, M.; Chin, H.F.; Lund, F.W.; Maxfield, F.R.; Breslow, J.L. Stard4 knockdown in HepG2 cells disrupts cholesterol trafficking associated with the plasma membrane, ER, and ERC. J. Lipid Res. 2012, 53, 2716–2725. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, M.; Fairn, G.D. Molecular probes to visualize the location, organization and dynamics of lipids. J. Cell Sci. 2014, 127, 4801–4812. [Google Scholar] [CrossRef] [PubMed]
- Bornig, H.; Geyer, G. Staining of cholesterol with the fluorescent antibiotic “filipin”. Acta Histochem. 1974, 50, 110–115. [Google Scholar] [PubMed]
- Li, Z.; Mintzer, E.; Bittman, R. First synthesis of free cholesterol-bodipy conjugates. J. Org. Chem. 2006, 71, 1718–1721. [Google Scholar] [CrossRef] [PubMed]
- Holtta-Vuori, M.; Uronen, R.L.; Repakova, J.; Salonen, E.; Vattulainen, I.; Panula, P.; Li, Z.; Bittman, R.; Ikonen, E. Bodipy-cholesterol: A new tool to visualize sterol trafficking in living cells and organisms. Traffic 2008, 9, 1839–1849. [Google Scholar] [CrossRef] [PubMed]
- Solanko, L.M.; Honigmann, A.; Midtiby, H.S.; Lund, F.W.; Brewer, J.R.; Dekaris, V.; Bittman, R.; Eggeling, C.; Wustner, D. Membrane orientation and lateral diffusion of bodipy-cholesterol as a function of probe structure. Biophys. J. 2013, 105, 2082–2092. [Google Scholar] [CrossRef] [PubMed]
- Heuck, A.P.; Hotze, E.M.; Tweten, R.K.; Johnson, A.E. Mechanism of membrane insertion of a multimeric β-barrel protein: Perfringolysin O creates a pore using ordered and coupled conformational changes. Mol. Cell 2000, 6, 1233–1242. [Google Scholar] [CrossRef]
- Heuck, A.P.; Tweten, R.K.; Johnson, A.E. Assembly and topography of the prepore complex in cholesterol-dependent cytolysins. J. Biol. Chem. 2003, 278, 31218–31225. [Google Scholar] [CrossRef] [PubMed]
- Shimada, Y.; Maruya, M.; Iwashita, S.; Ohno-Iwashita, Y. The C-terminal domain of Perfringolysin O is an essential cholesterol-binding unit targeting to cholesterol-rich microdomains. Eur. J. Biochem. 2002, 269, 6195–6203. [Google Scholar] [CrossRef] [PubMed]
- Kacprzyk-Stokowiec, A.; Kulma, M.; Traczyk, G.; Kwiatkowska, K.; Sobota, A.; Dadlez, M. Crucial role of Perfringolysin O D1 domain in orchestrating structural transitions leading to membrane-perforating pores: A hydrogen-deuterium exchange study. J. Biol. Chem. 2014, 289, 28738–28752. [Google Scholar] [CrossRef] [PubMed]
- Farrand, A.J.; LaChapelle, S.; Hotze, E.M.; Johnson, A.E.; Tweten, R.K. Only two amino acids are essential for cytolytic toxin recognition of cholesterol at the membrane surface. Proc. Natl. Acad Sci. USA 2010, 107, 4341–4346. [Google Scholar] [CrossRef] [PubMed]
- Soltani, C.E.; Hotze, E.M.; Johnson, A.E.; Tweten, R.K. Structural elements of the cholesterol-dependent cytolysins that are responsible for their cholesterol-sensitive membrane interactions. Proc. Natl. Acad Sci. USA 2007, 104, 20226–20231. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Sekino-Suzuki, N.; Mitsui, K.; Ohno-Iwashita, Y. Contribution of tryptophan residues to the structural changes in Perfringolysin O during interaction with liposomal membranes. J. Biochem. 1998, 123, 1145–1155. [Google Scholar] [CrossRef] [PubMed]
- Dowd, K.J.; Farrand, A.J.; Tweten, R.K. The cholesterol-dependent cytolysin signature motif: A critical element in the allosteric pathway that couples membrane binding to pore assembly. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef]
- Moe, P.C.; Heuck, A.P. Phospholipid hydrolysis caused by Clostridium perfringens α-toxin facilitates the targeting of Perfringolysin O to membrane bilayers. Biochemistry 2010, 49, 9498–9507. [Google Scholar] [CrossRef] [PubMed]
- Mobius, W.; van Donselaar, E.; Ohno-Iwashita, Y.; Shimada, Y.; Heijnen, H.F.; Slot, J.W.; Geuze, H.J. Recycling compartments and the internal vesicles of multivesicular bodies harbor most of the cholesterol found in the endocytic pathway. Traffic 2003, 4, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Abe, M.; Dedecker, P.; Makino, A.; Rocha, S.; Ohno-Iwashita, Y.; Hofkens, J.; Kobayashic, T.; Miyawakia, A. Fluorescent probes for superresolution imaging of lipid domains on the plasma membrane. Chem. Sci. 2011, 2, 1548–1553. [Google Scholar] [CrossRef]
- Das, A.; Goldstein, J.L.; Anderson, D.D.; Brown, M.S.; Radhakrishnan, A. Use of mutant 125I-Perfringolysin O to probe transport and organization of cholesterol in membranes of animal cells. Proc. Natl. Acad. Sci. USA 2013, 110, 10580–10585. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Brown, M.S.; Anderson, D.D.; Goldstein, J.L.; Radhakrishnan, A. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. Elife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Morita, I.; Fukuda, M.; Murota, S.; Ando, S.; Ohno-Iwashita, Y. A biotinylated Perfringolysin O derivative: A new probe for detection of cell surface cholesterol. Biochim. Biophys. Acta 1997, 1327, 222–230. [Google Scholar] [CrossRef]
- Mobius, W.; Ohno-Iwashita, Y.; van Donselaar, E.G.; Oorschot, V.M.; Shimada, Y.; Fujimoto, T.; Heijnen, H.F.; Geuze, H.J.; Slot, J.W. Immunoelectron microscopic localization of cholesterol using biotinylated and non-cytolytic Perfringolysin O. J. Histochem. Cytochem. 2002, 50, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Ohno-Iwashita, Y.; Iwamoto, M.; Mitsui, K.; Kawasaki, H.; Ando, S. Cold-labile hemolysin produced by limited proteolysis of θ-toxin from Clostridium perfringens. Biochemistry 1986, 25, 6048–6053. [Google Scholar] [CrossRef] [PubMed]
- Ohno-Iwashita, Y.; Iwamoto, M.; Ando, S.; Mitsui, K.; Iwashita, S. A modified theta-toxin produced by limited proteolysis and methylation: A probe for the functional study of membrane cholesterol. Biochim. Biophys. Acta 1990, 1023, 441–448. [Google Scholar] [CrossRef]
- Ohno-Iwashita, Y.; Iwamoto, M.; Mitsui, K.; Ando, S.; Iwashita, S. A cytolysin, θ-toxin, preferentially binds to membrane cholesterol surrounded by phospholipids with 18-carbon hydrocarbon chains in cholesterol-rich region. J. Biochem. 1991, 110, 369–375. [Google Scholar] [PubMed]
- Fisher, H.W.; Cooper, T.W. Electron microscope studies of the microvilli of HeLa cells. J. Cell Biol. 1967, 34, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Roper, K.; Corbeil, D.; Huttner, W.B. Retention of prominin in microvilli reveals distinct cholesterol-based lipid micro-domains in the apical plasma membrane. Nat. Cell Biol. 2000, 2, 582–592. [Google Scholar] [PubMed]
- Parton, R.G.; Hancock, J.F. Lipid rafts and plasma membrane microorganization: Insights from Ras. Trends Cell Biol. 2004, 14, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Carquin, M.; Conrard, L.; Pollet, H.; van der Smissen, P.; Cominelli, A.; Veiga-da-Cunha, M.; Courtoy, P.J.; Tyteca, D. Cholesterol segregates into submicrometric domains at the living erythrocyte membrane: Evidence and regulation. Cell. Mol. Life Sci. 2015, 72, 4633–4651. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.G. The use and abuse of filipin to localize cholesterol in membranes. Cell Biol. Int. Rep. 1984, 8, 519–535. [Google Scholar] [CrossRef]
- Ohno-Iwashita, Y.; Shimada, Y.; Hayashi, M.; Iwamoto, M.; Iwashita, S.; Inomata, M. Cholesterol-binding toxins and anti-cholesterol antibodies as structural probes for cholesterol localization. Subcellular Biochem. 2010, 51, 597–621. [Google Scholar]
- Saito, K.; Nishijima, M.; Kuge, O. Genetic evidence that phosphatidylserine synthase II catalyzes the conversion of phosphatidylethanolamine to phosphatidylserine in chinese hamster ovary cells. J. Biol. Chem. 1998, 273, 17199–17205. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.J.; Park, J.H.; Piggott, A.M.; Salim, A.A.; Gorfe, A.A.; Parton, R.G.; Capon, R.J.; Lacey, E.; Hancock, J.F. Staurosporines disrupt phosphatidylserine trafficking and mislocalize Ras proteins. J. Biol. Chem. 2012, 287, 43573–43584. [Google Scholar] [CrossRef] [PubMed]
- Raghupathy, R.; Anilkumar, A.A.; Polley, A.; Singh, P.P.; Yadav, M.; Johnson, C.; Suryawanshi, S.; Saikam, V.; Sawant, S.D.; Panda, A.; et al. Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell 2015, 161, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Yeung, T.; Gilbert, G.E.; Shi, J.; Silvius, J.; Kapus, A.; Grinstein, S. Membrane phosphatidylserine regulates surface charge and protein localization. Science 2008, 319, 210–213. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.J.; van der Hoeven, D.; Zhou, Y.; Maekawa, M.; Ma, X.; Chen, W.; Fairn, G.D.; Hancock, J.F. Inhibition of acid sphingomyelinase depletes cellular phosphatidylserine and mislocalizes K-Ras from the plasma membrane. Mol. Cell. Biol. 2015, 36, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Gulbins, E.; Palmada, M.; Reichel, M.; Luth, A.; Bohmer, C.; Amato, D.; Muller, C.P.; Tischbirek, C.H.; Groemer, T.W.; Tabatabai, G.; et al. Acid sphingomyelinase-ceramide system mediates effects of antidepressant drugs. Nat. Med. 2013, 19, 934–938. [Google Scholar] [CrossRef] [PubMed]
- Kornhuber, J.; Tripal, P.; Reichel, M.; Terfloth, L.; Bleich, S.; Wiltfang, J.; Gulbins, E. Identification of new functional inhibitors of acid sphingomyelinase using a structure-property-activity relation model. J. Med. Chem. 2008, 51, 219–237. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liang, H.; Rodkey, T.; Ariotti, N.; Parton, R.G.; Hancock, J.F. Signal integration by lipid-mediated spatial cross talk between Ras nanoclusters. Mol. Cell. Biol. 2014, 34, 862–876. [Google Scholar] [CrossRef] [PubMed]
- Hiramoto-Yamaki, N.; Tanaka, K.A.; Suzuki, K.G.; Hirosawa, K.M.; Miyahara, M.S.; Kalay, Z.; Tanaka, K.; Kasai, R.S.; Kusumi, A.; Fujiwara, T.K. Ultrafast diffusion of a fluorescent cholesterol analog in compartmentalized plasma membranes. Traffic 2014, 15, 583–612. [Google Scholar] [CrossRef] [PubMed]
- Saka, S.K.; Honigmann, A.; Eggeling, C.; Hell, S.W.; Lang, T.; Rizzoli, S.O. Multi-protein assemblies underlie the mesoscale organization of the plasma membrane. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, J.J.; Tweten, R.K.; Johnson, A.E.; Heuck, A.P. Cholesterol exposure at the membrane surface is necessary and sufficient to trigger Perfringolysin O binding. Biochemistry 2009, 48, 3977–3987. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, A.; Radhakrishnan, A. Accessibility of cholesterol in endoplasmic reticulum membranes and activation of SREBP-2 switch abruptly at a common cholesterol threshold. J. Biol. Chem. 2010, 285, 29480–29490. [Google Scholar] [CrossRef] [PubMed]



| Name | Structure | Mutations | Type | Temperature | Localization | Devices for Detection | Live-Imaging |
|---|---|---|---|---|---|---|---|
| PFO* | Full length | Y181A/C459A | 125I-labeled recombinant proteins | 4 °C | PM (exofacial) | Scintillation counter | No |
| BCθ | Full length | No | Biotinylated recombinant proteins | RT | PM, endosomes | Electron microscopy/Fluorescence microscopy | No/Yes (with Avidin) |
| D4 | Domain 4 | No | Fluorophore-labeled recombinant proteins | RT or 4 °C | PM (exofacial) | Fluorescence microscopy, PALM Flow cytometry | Yes |
| D4H | Domain 4 | D434S | Vector-based (with fluorophore) | 37 °C | PM, endosomes (cytosolic) | Fluorescence microscopy | Yes |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maekawa, M.; Yang, Y.; Fairn, G.D. Perfringolysin O Theta Toxin as a Tool to Monitor the Distribution and Inhomogeneity of Cholesterol in Cellular Membranes. Toxins 2016, 8, 67. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins8030067
Maekawa M, Yang Y, Fairn GD. Perfringolysin O Theta Toxin as a Tool to Monitor the Distribution and Inhomogeneity of Cholesterol in Cellular Membranes. Toxins. 2016; 8(3):67. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins8030067
Chicago/Turabian StyleMaekawa, Masashi, Yanbo Yang, and Gregory D. Fairn. 2016. "Perfringolysin O Theta Toxin as a Tool to Monitor the Distribution and Inhomogeneity of Cholesterol in Cellular Membranes" Toxins 8, no. 3: 67. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins8030067