Probing the Characterization of the Interaction of Aflatoxins B1 and G1 with Calf Thymus DNA In Vitro
Abstract
:1. Introduction
2. Results
2.1. Dialysis Assay
2.2. Fluorescence and Resonance Light Scattering (RLS) Spectra
2.2.1. Fluorescence Studies
2.2.2. RLS Spectral Analysis
2.3. Competitive Displacement Assays
2.4. UV–Visible Spectroscopy
2.5. Viscosity Measurements
2.6. DNA Melting Assay
2.7. Effect of Ionic Intensity
2.8. Effects of Aflatoxins on Native or Denatured DNA
2.9. Circular Dichroism Spectral Measurement
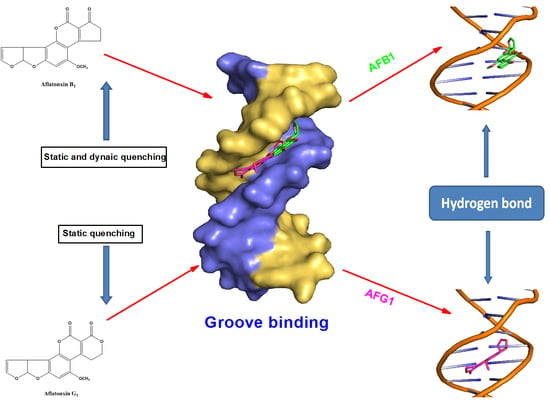
2.10. Molecular Docking Studies
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Instrumentation
4.3. Dialysis Assays
4.4. Fluorescence and Resonance Light Scattering (RLS) Spectra
4.5. Competitive Displacement Assays
4.6. UV–Visible Spectroscopy
4.7. Viscosity Measurements
4.8. DNA Melting Assay
4.9. Effect of Ionic Intensity
4.10. Interaction with Single-Stranded ctDNA (ss-ctDNA) and Double-Stranded ctDNA (ds-ctDNA)
4.11. Circular Dichroism (CD) Spectroscopy
4.12. Molecular Docking
4.13. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Richard, E.; Heutte, N.; Sage, L.; Pottier, D.; Bouchart, V.; Lebailly, P.; Garon, D. Toxigenic fungi and mycotoxins in mature corn silage. Food Chem. Toxicol. 2007, 45, 2420–2425. [Google Scholar] [CrossRef] [PubMed]
- Herebian, D.; Zuehlke, S.; Lamshoeft, M.; Spiteller, M. Multi-mycotoxin analysis in complex biological matrices using LC-ESI/MS: Experimental study using triple stage quadrupole and LTQ-orbitrap. J. Sep. Sci. 2009, 32, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Schenzel, J.; Forrer, H.R.; Vogelgsang, S.; Hungerbuehler, K.; Bucheli, T.D. Mycotoxins in the environment: I. Production and emission from an agricultural test field. Environ. Sci. Technol. 2012, 46, 13067–13075. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Chen, X.; Wang, Y.; Zheng, X.; Li, C.M. Aptamer based fluorescence recovery assay for aflatoxin B1 using a quencher system composed of quantum dots and graphene oxide. Microchim. Acta 2015, 182, 571–578. [Google Scholar] [CrossRef]
- Ivanova, B.; Spiteller, M. Raman spectroscopic and mass spectrometric determination of aflatoxins. Food Anal. Methods 2014, 7, 242–256. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer (IARC). IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; IARC Press: Lyon, France, 2002; pp. 171–230. [Google Scholar]
- Eaton, D.L.; Gallagher, E.P. Mechanism of aflatoxin carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 1994, 34, 135–172. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Wei, M.; Kokot, S. Electrochemical and spectroscopic study on the interaction between isoprenaline and DNA using multivariate curve resolution-alternating least squares. Int. J. Biol. Macromol. 2011, 49, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, G.; Li, Y.; Hu, Y. Probing the binding of insecticide permethrin to calf thymus DNA by spectroscopic techniques merging with chemometrics method. J. Agric. Food Chem. 2013, 61, 2638–2647. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Huang, Y.; Zhang, Y.; Song, L.; Qiao, Y.; Xu, X.; Wang, C. Spectroscopic and molecular modeling methods to study the interaction between naphthalimide-polyamine conjugates and DNA. J. Photochem. Photobiol. B 2016, 158, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Mehta, J.; Rouah-Martin, E.; Van Dorst, B.; Maes, B.; Herrebout, W.; Scippo, M.L.; Dardenne, F.; Blust, R.; Robbens, J. Selection and characterization of pcb-binding DNA aptamers. Anal. Chem. 2012, 84, 1669–1676. [Google Scholar] [CrossRef] [PubMed]
- Hendry, L.B.; Mahesh, V.B.; Bransome, E.D.; Ewing, D.E. Small molecule intercalation with double stranded DNA: Implications for normal gene regulation and for predicting the biological efficacy and genotoxicity of drugs and other chemicals. Mutat. Res. 2007, 623, 53–71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Ding, L. Interaction between tryptophan-vanillin schiff base and herring sperm DNA. J. Serb. Chem. Soc. 2010, 75, 1191–1201. [Google Scholar] [CrossRef]
- Croy, R.G.; Essigmann, J.M.; Reinhold, V.N.; Wogan, G.N. Identification of principal aflatoxin B1-DNA adduct formed invivo in rat-liver. Proc. Natl. Acad. Sci. USA 1978, 75, 1745–1749. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.S.; Groopman, J.D. DNA damage by mycotoxins. Mutat. Res. 1999, 424, 167–181. [Google Scholar] [CrossRef]
- Shahabadi, N.; Heidari, L. Synthesis, characterization and multi-spectroscopic DNA interaction studies of a new platinum complex containing the drug metformin. Spectrochim. Acta A 2014, 128, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, G.; Tao, M. Binding properties of herbicide chlorpropham to DNA: Spectroscopic, chemometrics and modeling investigations. J. Photochem. Photobiol. B 2014, 138, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Shahabadi, N.; Kashanian, S.; Darabi, F. DNA binding and DNA cleavage studies of a water soluble cobalt(II) complex containing dinitrogen schiff base ligand: The effect of metal on the mode of binding. Eur. J. Med. Chem. 2010, 45, 4239–4245. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of fluorescence spectroscopy. In Fluorophores, 3rd ed.; Springer: New York, NY, USA, 2008. [Google Scholar]
- Lakowicz, J.R.; Weber, G. Quenching of fluorescence by oxygen-probe for structural fluctuations in macromolecules. Biochemistry 1973, 12, 4161–4170. [Google Scholar] [CrossRef] [PubMed]
- Sneharani, A.H.; Karakkat, J.V.; Singh, S.A.; Rao, A.G.A. Interaction of curcumin with beta-lactoglobulin-stability, spectroscopic analysis, and molecular modeling of the complex. J. Agric. Food Chem. 2010, 58, 11130–11139. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.M.; Xi, X.L.; Yang, P. Study of the interaction of cephalosporin class medicine with albumin by fluorescence enhancement and fluorescence quenching theories. Chin. J. Chem. 2006, 24, 642–648. [Google Scholar] [CrossRef]
- Hu, Y.J.; Liu, Y.; Xiao, X.H. Investigation of the interaction between berberine and human serum albumin. Biomacromolecules 2009, 10, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Darwish, S.M.; Abu Sharkh, S.E.; Abu Teir, M.M.; Makharza, S.A.; Abu-Hadid, M.M. Spectroscopic investigations of pentobarbital interaction with human serum albumin. J. Mol. Struct. 2010, 963, 122–129. [Google Scholar] [CrossRef]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions-forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, X.; Fu, R.; Luo, W.A.; Li, Y.; Zhang, M. Kinetics of phase separation in polymer blends revealed by resonance light scattering spectroscopy. Phys. Chem. Chem. Phys. 2010, 12, 2238–2245. [Google Scholar] [CrossRef] [PubMed]
- He, Y.Q.; Liu, S.P.; Kong, L.; Liu, Z.F. A study on the sizes and concentrations of gold nanoparticles by spectra of absorption, resonance rayleigh scattering and resonance non-linear scattering. Spectrochim. Acta A 2005, 61, 2861–2866. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Fu, P.; Wang, L.; Hu, M. Molecular spectroscopic studies of farrerol interaction with calf thymus DNA. J. Agric. Food Chem. 2011, 59, 8944–8952. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.U.; Sarwar, T.; Ishqi, H.M.; Husain, M.A.; Hasan, Z.; Tabish, M. Deciphering the interactions between chlorambucil and calf thymus DNA: A multi-spectroscopic and molecular docking study. Arch. Biochem. Biophys. 2015, 566, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Baguley, B.C.; Lebret, M. Quenching of DNA-ethidium fluorescence by amsacrine and other antitumor agents-a possible electron-transfer effect. Biochemistry 1984, 23, 937–943. [Google Scholar] [CrossRef] [PubMed]
- Loontiens, F.G.; Regenfuss, P.; Zechel, A.; Dumortier, L.; Clegg, R.M. Binding characteristics of hoechst-33258 with calf thymus DNA, poly d(AT), and d(CCGGAATTCCGG)-multiple stoichiometries and determination of tight-binding with a wide spectrum of site affinities. Biochemistry 1990, 29, 9029–9039. [Google Scholar] [CrossRef] [PubMed]
- Skyrianou, K.C.; Psycharis, V.; Raptopoulou, C.P.; Kessissoglou, D.P.; Psomas, G. Nickel-quinolones interaction part 4-structure and biological evaluation of nickel (II)-enrofloxacin complexes compared to zinc(II) analogues. J. Inorg. Biochem. 2011, 105, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Vujcic, M.T.; Tufegdzic, S.; Novakovic, I.; Djikanovic, D.; Gasic, M.J.; Sladic, D. Studies on the interactions of bioactive quinone avarone and its methylamino derivatives with calf thymus DNA. Int. J. Biol. Macromol. 2013, 62, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Aramesh-Boroujeni, Z.; Khorasani-Motlagh, M.; Noroozifar, M. Multispectroscopic DNA-binding studies of a terbium(III) complex containing 2,2’-bipyridine ligand. J. Biomol. Struct. Dyn. 2016, 34, 414–426. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.N.; Zhan, C.X. Studies on the interaction of DNA and water-soluble polymeric schiff base-nickel complexes. J. Appl. Polym. Sci. 2002, 84, 887–893. [Google Scholar] [CrossRef]
- Devi, C.V.; Singh, N.R. Absorption spectroscopic probe to investigate the interaction between Nd(III) and calf-thymus DNA. Spectrochim. Acta A 2011, 78, 1180–1186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wang, L.; Zhou, X.; Li, Y.; Gong, D. Binding characteristics of sodium saccharin with calf thymus DNA in vitro. J. Agric. Food Chem. 2014, 62, 991–1000. [Google Scholar] [CrossRef] [PubMed]
- Kashanian, S.; Khodaei, M.M.; Kheirdoosh, F. In vitro DNA binding studies of aspartame, an artificial sweetener. J. Photochem. Photobiol. B 2013, 120, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Charak, S.; Shandilya, M.; Tyagi, G.; Mehrotra, R. Spectroscopic and molecular docking studies on chlorambucil interaction with DNA. Int. J. Biol. Macromol. 2012, 51, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Kashanian, S.; Dolatabadi, J.E.N. DNA binding studies of 2-tert-butylhydroquinone (TBHQ) food additive. Food Chem. 2009, 116, 743–747. [Google Scholar] [CrossRef]
- Bera, R.; Sahoo, B.K.; Ghosh, K.S.; Dasgupta, S. Studies on the interaction of isoxazolcurcumin with calf thymus DNA. Int. J. Biol. Macromol. 2008, 42, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Huo, R.; Hui, G.; Lv, X.; Jin, J.; Zhang, G.; Xing, W. Study on the interaction between aglycon of daunorubicin and calf thymus DNA by spectroscopy. J. Mol. Struct. 2011, 1001, 104–110. [Google Scholar] [CrossRef]
- Jana, B.; Senapati, S.; Ghosh, D.; Bose, D.; Chattopadhyay, N. Spectroscopic exploration of mode of binding of ctDNA with 3-hydroxyflavone: A contrast to the mode of binding with flavonoids having additional hydroxyl groups. J. Chem. Phys. B 2012, 116, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.Y.; Xiang, Y.L.; Wu, Y.M.; Xie, F.Y. Study of interaction of a fluorescent probe with DNA. J. Lumin. 2009, 129, 1286–1291. [Google Scholar] [CrossRef]
- Cai, C.; Chen, X.; Ge, F. Analysis of interaction between tamoxifen and ctDNA in vitro by multi-spectroscopic methods. Spectrochim. Acta A 2010, 76, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Xiang, G.; Bai, Y. Interaction of paraquat with calf thymus DNA: A terbium(III) luminescent probe and multispectral study. J. Agric. Food Chem. 2010, 58, 5257–5262. [Google Scholar] [CrossRef] [PubMed]
- Shahabadi, N.; Moghadam, N.H. Determining the mode of interaction of calf thymus DNA with the drug sumatriptan using voltammetric and spectroscopic techniques. Spectrochim. Acta A 2012, 99, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, G.; Charak, S.; Mehrotra, R. Binding of an indole alkaloid, vinblastine to double stranded DNA: A spectroscopic insight in to nature and strength of interaction. J. Photochem. Photobiol. B 2012, 108, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.M.; Liu, J.; Chen, J.C.; Shi, S.; Tan, C.P.; Zheng, K.C.; Ji, L.N. Experimental and theoretical studies on the DNA-binding and spectral properties of water-soluble complex [Ru(Meim)(4)(dpq)]2+. J. Mol. Struct. 2008, 881, 156–166. [Google Scholar] [CrossRef]
- Maheswari, P.U.; Palaniandavar, M. DNA binding and cleavage properties of certain tetrammine ruthenium(II) complexes of modified 1,10-phenanthrolines-effect of hydrogen-bonding on DNA-binding affinity. J. Inorg. Biochem. 2004, 98, 219–230. [Google Scholar] [CrossRef]
- Tao, M.; Zhang, G.; Pan, J.; Xiong, C. Deciphering the groove binding modes of tau-fluvalinate and flumethrin with calf thymus DNA. Spectrochim. Acta A 2016, 155, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, R.; Mahasenan, K.; Pavlovicz, R.; Li, C.; Tjarks, W. Carborane clusters in computational drug design: A comparative docking evaluation using autodock, flexX, glide, and surflex. J. Chem. Inf. Model. 2009, 49, 1581–1589. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.; Liu, R. Phenotypic characterization of the binding of tetracycline to human serum albumin. Biomacromolecules 2011, 12, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, B.; Spiteller, M. Structure and properties of camptothecin derivatives, their protonated forms, and model interaction with the topoisomerase I-DNA complex. Biopolymers 2012, 97, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Privalov, P.L.; Dragan, A.I.; Crane-Robinson, C.; Breslauer, K.J.; Remeta, D.P.; Minetti, C.A.S.A. What drives proteins into the major or minor grooves of DNA? J. Mol. Biol. 2007, 365, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, B.; Spiteller, M. Substituted benzo i phenanthridines as promising topoisomerase-i non-camptothecin targeting agents: An experimental and theoretical study. Med. Chem. Res. 2013, 22, 5204–5217. [Google Scholar] [CrossRef]
- Bi, S.; Zhao, T.; Wang, Y.; Zhou, H.; Pang, B.; Gu, T. Binding studies of terbutaline sulfate to calf thymus DNA using multispectroscopic and molecular docking techniques. Spectrochim. Acta A 2015, 150, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Song, G.W.; He, Y.; Cai, Z.X. The interaction between levofloxacine hydrochloride and DNA mediated by Cu2+. J. Fluoresc. 2004, 14, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Husain, M.A.; Sarwar, T.; Rehman, S.U.; Ishqi, H.M.; Tabish, M. Ibuprofen causes photocleavage through ros generation and intercalates with DNA: A combined biophysical and molecular docking approach. Phys. Chem. Chem. Phys. 2015, 17, 13837–13850. [Google Scholar] [CrossRef] [PubMed]














| Aflatoxins | T (K) | Ksv a (L·mol−1) | R b | Ka c (L·mol−1) | R d |
|---|---|---|---|---|---|
| AFB1 | 298 | 1.24 × 104 | 0.9938 | 4.12 × 103 | 0.9921 |
| 303 | 2.31 × 104 | 0.9959 | 4.97 × 103 | 0.9901 | |
| 308 | 3.22 × 104 | 0.9902 | 5.48 × 103 | 0.9881 | |
| AFG1 | 298 | 8.31 × 104 | 0.9912 | 1.89 × 104 | 0.9865 |
| 303 | 7.32 × 104 | 0.9887 | 1.27 × 104 | 0.9908 | |
| 308 | 0.55 × 104 | 0.9908 | 9.82 × 103 | 0.9918 |
| Systems | ΔH a (kJ·mol−1) | ΔS b (J·mol−1·K−1) | ΔG c (kJ·mol−1) |
|---|---|---|---|
| AFB1-ctDNA | −24.45 | 17.88 | −31.47 |
| −31.98 | |||
| −32.56 | |||
| AFG1-ctDNA | −27.54 | −19.34 | −36.32 |
| −35.97 | |||
| −35.65 |
| Complex | Intermolecular Energy (kcal·mol−1) | Internal Energy (kcal·mol−1) | Torsional Energy (kcal·mol−1) | Unbound Extended Energy (kcal·mol−1) | Lowest Binding Energy (kcal·mol−1) | Electrostatic Energy (kcal·mol−1) |
|---|---|---|---|---|---|---|
| AFB1-DNA | −8.08 | −0.12 | 0.30 | −0.12 | −7.78 | −0.46 |
| AFG1-DNA | −8.59 | −0.09 | 0.30 | −0.09 | −8.29 | 0.04 |
| Small Molecular | Strong Hydrogen Bonds | Moderate Hydrogen Bonds | Weak Hydrogen Bonds |
|---|---|---|---|
| AFB1 | 2.6 Å (DG-16) 2.8 Å (DA-17) 2.9 Å (DA-17) | ||
| AFG1 | 2.9 Å (DT-7) 2.9 Å (DT-8) | 3.1 Å (DT-19) 3.2 Å (DA-18) 3.4 Å (DT-19) | 3.6 Å (DT-20) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, L.; Wang, J.; Zhang, Y. Probing the Characterization of the Interaction of Aflatoxins B1 and G1 with Calf Thymus DNA In Vitro. Toxins 2017, 9, 209. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins9070209
Ma L, Wang J, Zhang Y. Probing the Characterization of the Interaction of Aflatoxins B1 and G1 with Calf Thymus DNA In Vitro. Toxins. 2017; 9(7):209. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins9070209
Chicago/Turabian StyleMa, Liang, Jiaman Wang, and Yuhao Zhang. 2017. "Probing the Characterization of the Interaction of Aflatoxins B1 and G1 with Calf Thymus DNA In Vitro" Toxins 9, no. 7: 209. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins9070209