Roles of Histone Deacetylases and Inhibitors in Anticancer Therapy
Abstract
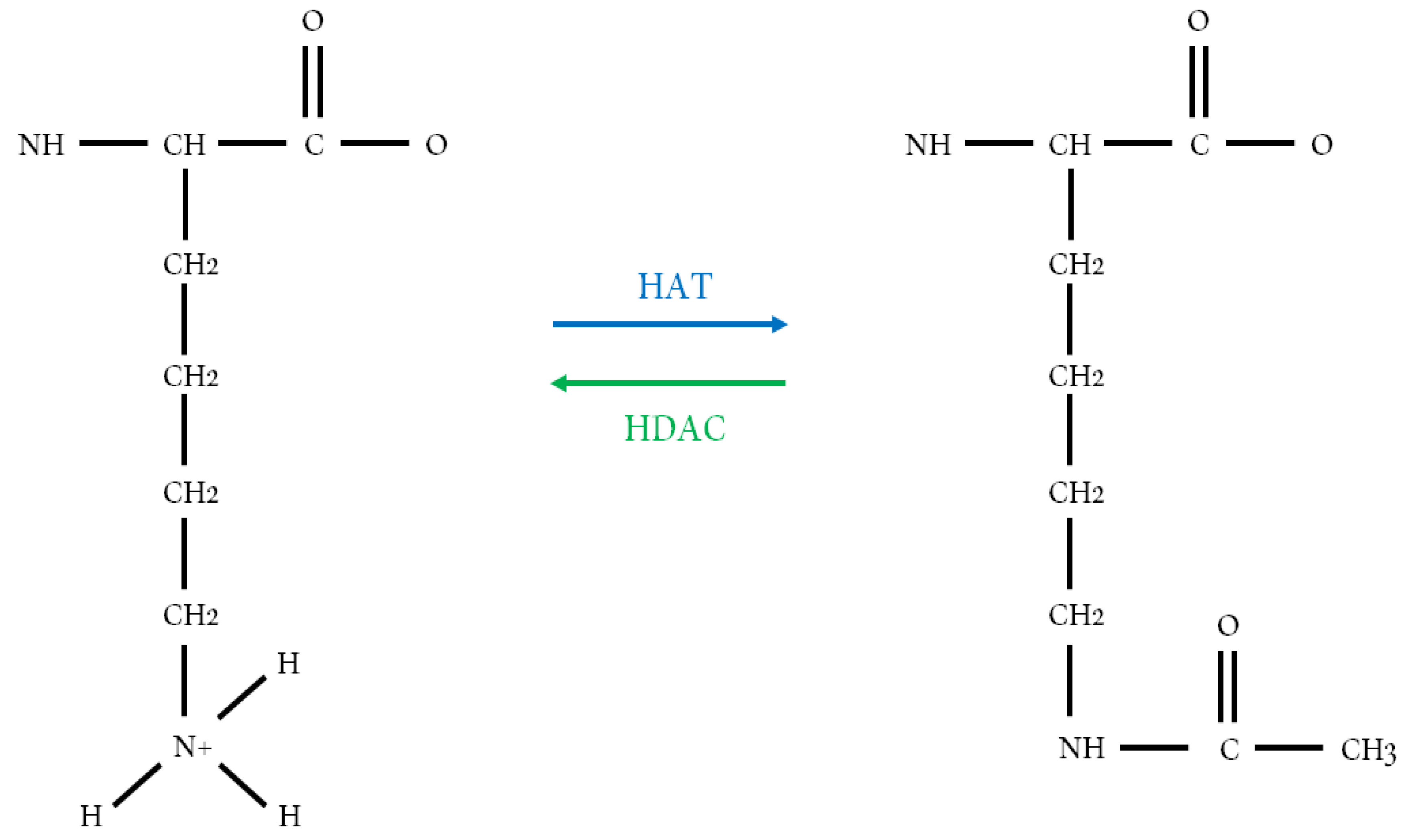
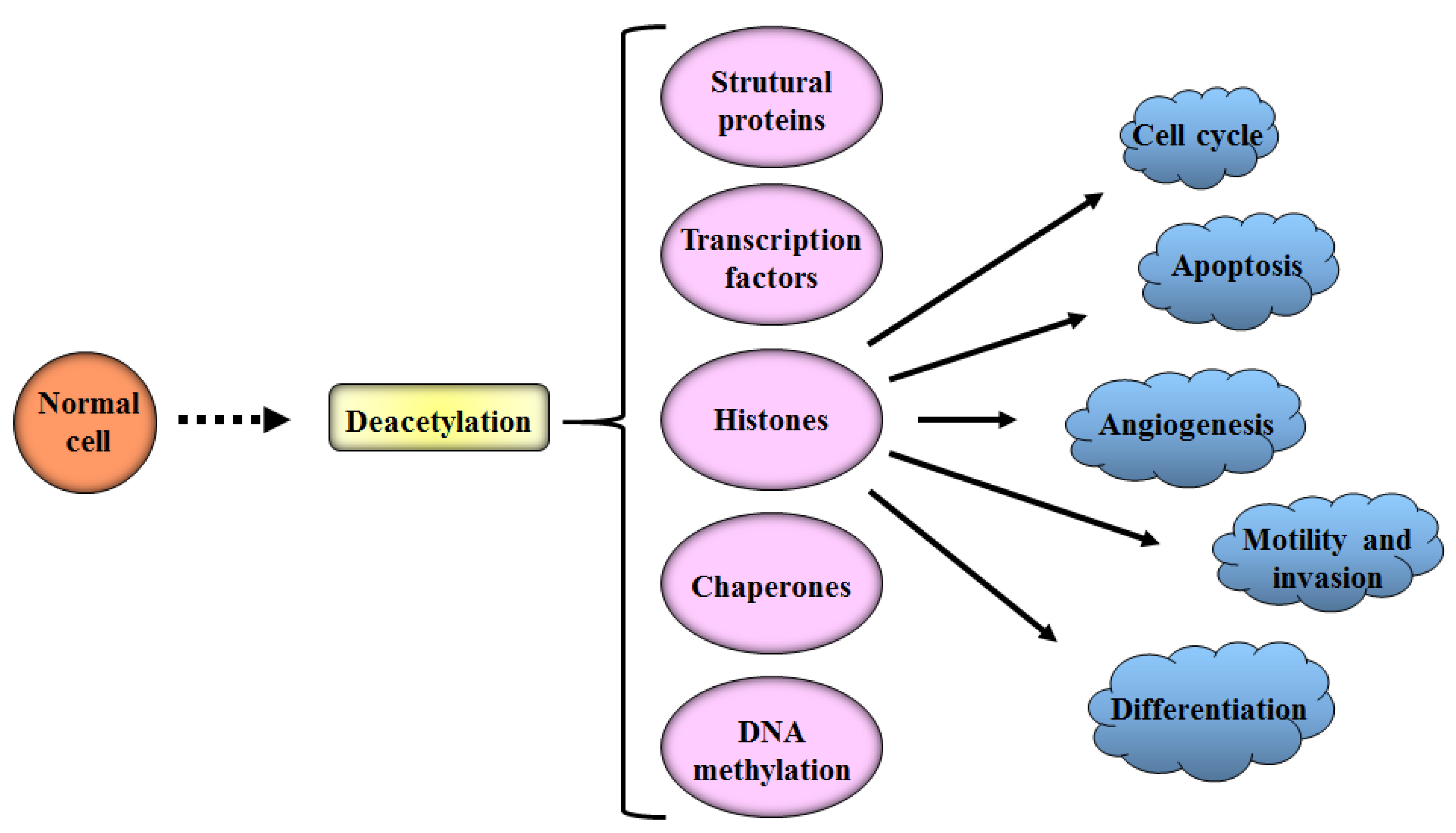
:1. Introduction
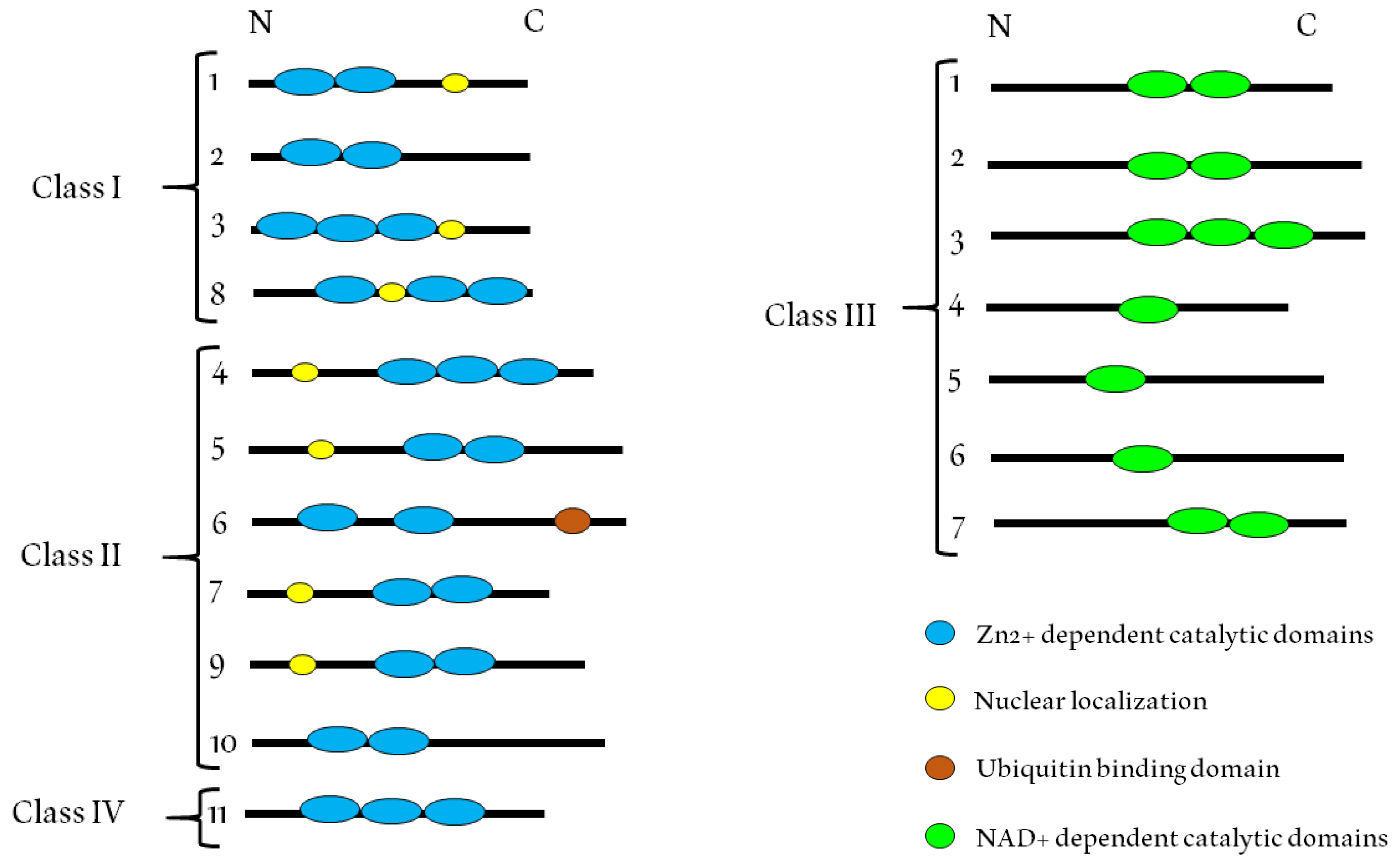
2. Class I HDACs
2.1. The HDAC1/2 Functional Complex
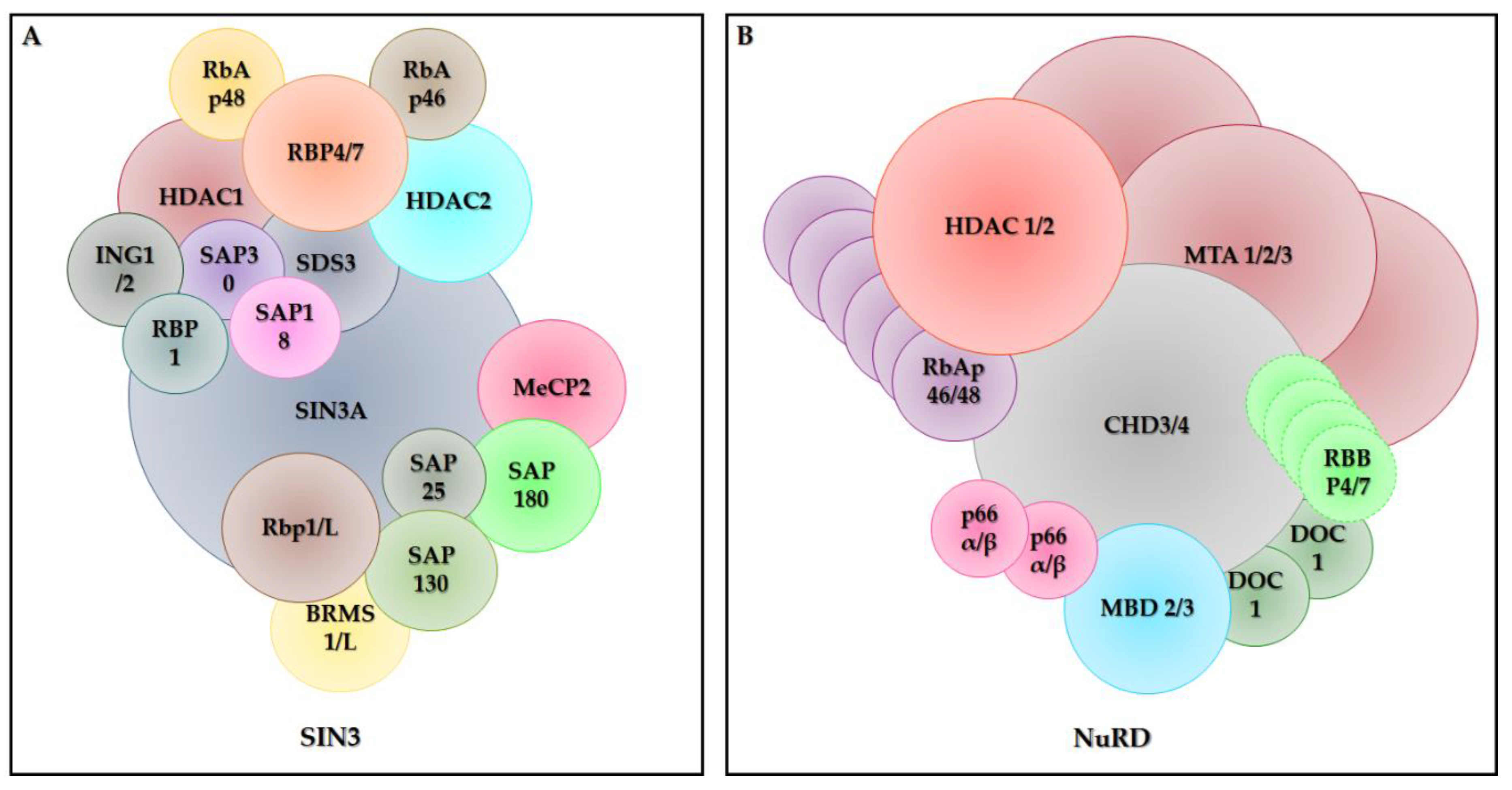
2.2. Sin3A Complex
2.3. NuRD Complex
3. Class II HDACs
4. Class IV HDACs
5. Participation of Transcription Factors in the Mechanism of Regulation by HDACs
6. Inhibitors of HDACs
7. Alternative Inhibitors
7.1. Chalcones
7.2. Curcuminoids
8. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Cheung, W.L.; Briggs, S.D.; Allis, C.D. Acetylation and chromosomal functions. Curr. Opin. Cell Boil. 2000, 12, 326–333. [Google Scholar] [CrossRef]
- Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 1997, 389, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Gundersen, G.G.; Worman, H.J. Nuclear positioning. Cell 2013, 152, 1376–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornberg, R.D. Chromatin Structure: A Repeating Unit of Histones and DNA. Science 1974, 184, 868–871. [Google Scholar] [CrossRef]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- De La Cruz, X.; Lois, S.; Sánchez-Molina, S.; Martínez-Balbás, M.A. Do protein motifs read the histone code? BioEssays 2005, 27, 164–175. [Google Scholar] [CrossRef] [Green Version]
- Woodcock, C.L.; Skoultchi, A.I.; Fan, Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosom. Res. 2006, 14, 17–25. [Google Scholar] [CrossRef]
- Hansen, J.C. Conformational Dynamics of the Chromatin Fiber in Solution: Determinants, Mechanisms, and Functions. Annu. Rev. Biophys. Biomol. Struct. 2002, 31, 361–392. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacol. 2012, 38, 23–38. [Google Scholar] [CrossRef] [Green Version]
- Allfrey, V.G.; Mirsky, A.E.; Arnon, D.I.; Tsujimoto, H.Y.; McSwain, B.D. Structural Modifications of Histones and their Possible Role in the Regulation of RNA Synthesis. Sciebce 1964, 144, 559. [Google Scholar] [CrossRef]
- Wolffe, A.P.; Hayes, J.J. Chromatin disruption and modification. Nucleic Acids Res. 1999, 27, 711–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edmondson, D.G.; Smith, M.M.; Roth, S.Y. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996, 10, 1247–1259. [Google Scholar] [CrossRef] [Green Version]
- Zentner, G.E.; Henikoff, S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Boil. 2013, 20, 259–266. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Manning, L.R.; Russell, J.E.; Popowicz, A.M.; Manning, R.S.; Padovan, J.C.; Manning, J.M. Energetic Differences at the Subunit Interfaces of Normal Human Hemoglobins Correlate with Their Developmental Profile. Biochemistry 2009, 48, 7568–7574. [Google Scholar] [CrossRef] [Green Version]
- Rajendran, P.; Delage, B.; Dashwood, W.M.; Yu, T.-W.; Wuth, B.; Williams, D.E.; Ho, E.; Dashwood, R.H. Histone deacetylase turnover and recovery in sulforaphane-treated colon cancer cells: Competing actions of 14-3-3 and Pin1 in HDAC3/SMRT corepressor complex dissociation/reassembly. Mol. Cancer 2011, 10, 68. [Google Scholar] [CrossRef] [Green Version]
- Spange, S.; Wagner, T.; Heinzel, T.; Krämer, O. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Boil. 2009, 41, 185–198. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, Y.; Shi, X. Emerging roles of lysine methylation on non-histone proteins. Cell. Mol. Life Sci. 2015, 72, 4257–4272. [Google Scholar] [CrossRef]
- Abbass, S.A.; Hassan, H.A.; Mohamed, M.F.A.; Moustafa, G.A.I.; Abuo-Rahma, G.E.-D.A. Recent Prospectives of Anticancer Histone Deacetylase Inhibitors. J. Adv. Biomed. Pharm. Sci. 2019, 2, 135–151. [Google Scholar] [CrossRef] [Green Version]
- Iizuka, M.; Smith, M.M. Functional consequences of histone modifications. Curr. Opin. Genet. Dev. 2003, 13, 154–160. [Google Scholar] [CrossRef]
- Berger, S. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002, 12, 142–148. [Google Scholar] [CrossRef]
- Gibbon, S.; Prainsack, B.; Hilgartner, S.; Lamoreaux, J. (Eds.) Routledge Handbook of Genomics, Health and Society; Routledge: Abington, UK, 2018. [Google Scholar]
- Parthun, M.R. Hat1: The emerging cellular roles of a type B histone acetyltransferase. Oncogene 2007, 26, 5319–5328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polo, S.; Almouzni, G. Histone metabolic pathways and chromatin assembly factors as proliferation markers. Cancer Lett. 2005, 220, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Vidanes, G.M.; Bonilla, C.Y.; Toczyski, D.P. Complicated Tails: Histone Modifications and the DNA Damage Response. Cell 2005, 121, 973–976. [Google Scholar] [CrossRef] [Green Version]
- Hendrich, B.; Bickmore, W.A. Human diseases with underlying defects in chromatin structure and modification. Hum. Mol. Genet. 2001, 10, 2233–2242. [Google Scholar] [CrossRef] [Green Version]
- Cairns, B.R. Emerging roles for chromatin remodeling in cancer biology. Trends Cell Boil. 2001, 11, S15–S21. [Google Scholar] [CrossRef]
- Hake, S.B.; Xiao, A.; Allis, C.D. Linking the epigenetic ‘language’ of covalent histone modifications to cancer. Br. J. Cancer 2004, 90, 761–769. [Google Scholar] [CrossRef] [Green Version]
- Tamkun, J.W.; Deuring, R.; Scott, M.P.; Kissinger, M.; Pattatucci, A.M.; Kaufman, T.C.; Kennison, J.A. brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2SWI2. Cell 1992, 68, 561–572. [Google Scholar] [CrossRef]
- Gil, J.; Ramírez-Torres, A.; Guevara, S.E. Lysine acetylation and cancer: A proteomics perspective. J. Proteom. 2017, 150, 297–309. [Google Scholar] [CrossRef]
- Roth, S.Y.; Denu, J.M.; Allis, C.D. Histone acetyltransferases. Annu. Rev. Biochem. 2001, 70, 81–120. [Google Scholar] [CrossRef]
- Dekker, F.J.; Haisma, H.J. Histone acetyl transferases as emerging drug targets. Drug Discov. Today 2009, 14, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Clayton, A.L.; Hazzalin, C.A.; Mahadevan, L.C. Enhanced Histone Acetylation and Transcription: A Dynamic Perspective. Mol. Cell 2006, 23, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Paik, W.K.; Borun, T.W. The periodic synthesis of S-adenosylmethionine: Protein methyltransferases during the HeLa S-3 cell cycle. J. Boil. Chem. 1973, 248, 4194–4199. [Google Scholar]
- Wolffe, A.P.; Guschin, D. Review: Chromatin Structural Features and Targets That Regulate Transcription. J. Struct. Boil. 2000, 129, 102–122. [Google Scholar] [CrossRef] [Green Version]
- Wapenaar, H.; Dekker, F.J. Histone acetyltransferases: Challenges in targeting bi-substrate enzymes. Clin. Epigenetics 2016, 8, 59. [Google Scholar] [CrossRef] [Green Version]
- Burgess, R.J.; Zhang, Z. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Boil. 2013, 20, 14–22. [Google Scholar] [CrossRef] [Green Version]
- Haberland, M.; Montgomery, R.L.; Olson, E. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat. Rev. Genet. 2009, 10, 32–42. [Google Scholar] [CrossRef]
- De Ruijter, A.J.; Van Gennip, A.H.; Caron, H.N.; Kemp, S.; Van Kuilenburg, A.B. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003, 370, 737–749. [Google Scholar] [CrossRef]
- Inoue, A.; Fujimoto, D. Enzymatic deacetylation of histone. Biochem. Biophys. Res. Commun. 1969, 36, 146–150. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb. Perspect. Boil. 2014, 6, a018713. [Google Scholar] [CrossRef] [Green Version]
- Ropero, S.; Esteller, M. The role of histone deacetylases (HDACs) in human cancer. Mol. Oncol. 2007, 1, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Dokmanovic, M.; Marks, P.A. Prospects: Histone deacetylase inhibitors. J. Cell. Biochem. 2005, 96, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Rosato, R.R.; Grant, S. Histone deacetylase inhibitors: Insights into mechanisms of lethality. Expert Opin. Ther. Targets 2005, 9, 809–824. [Google Scholar] [CrossRef] [PubMed]
- Frew, A.J.; Johnstone, R.W.; Bolden, J.E. Enhancing the apoptotic and therapeutic effects of HDAC inhibitors. Cancer Lett. 2009, 280, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.N.; Zhang, G.; Hwa, Y.L.; Li, J.; Dowdy, S.C.; Jiang, S.-W. Nonhistone protein acetylation as cancer therapy targets. Expert Rev. Anticancer. Ther. 2010, 10, 935–954. [Google Scholar] [CrossRef] [Green Version]
- Lane, A.A.; Chabner, B.A. Histone Deacetylase Inhibitors in Cancer Therapy. J. Clin. Oncol. 2009, 27, 5459–5468. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.-Y.; Chen, C.-S.; Lin, S.-P.; Weng, J.-R.; Chen, C.-S. Targeting Histone Deacetylase in Cancer Therapy. Chemin 2006, 37, 397–413. [Google Scholar] [CrossRef]
- Lagger, G.; O’Carroll, D.; Rembold, M.; Khier, H.; Tischler, J.; Weitzer, G.; Schuettengruber, B.; Häuser, C.; Brunmeir, R.; Jenuwein, T.; et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002, 21, 2672–2681. [Google Scholar] [CrossRef] [Green Version]
- Vega, R.B.; Matsuda, K.; Oh, J.; Barbosa, A.C.; Yang, X.; Meadows, E.; McAnally, J.; Pomajzl, C.; Shelton, J.M.; Richardson, J.A.; et al. Histone Deacetylase 4 Controls Chondrocyte Hypertrophy during Skeletogenesis. Cell 2004, 119, 555–566. [Google Scholar] [CrossRef] [Green Version]
- Arnold, M.; Kim, Y.; Czubryt, M.P.; Phan, D.; McAnally, J.; Qi, X.; Shelton, J.M.; Richardson, J.A.; Bassel-Duby, R.; Olson, E. MEF2C Transcription Factor Controls Chondrocyte Hypertrophy and Bone Development. Dev. Cell 2007, 12, 377–389. [Google Scholar] [CrossRef] [Green Version]
- Rettig, I.; Koeneke, E.; Trippel, F.; Mueller, W.C.; Burhenne, J.; Kopp-Schneider, A.; Fabian, J.; Schober, A.; Fernekorn, U.; Von Deimling, A.; et al. Selective inhibition of HDAC8 decreases neuroblastoma growth in vitro and in vivo and enhances retinoic acid-mediated differentiation. Cell Death Dis. 2015, 6, e1657. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yamashita, H.; Toyama, T.; Sugiura, H.; Ando, Y.; Mita, K.; Hamaguchi, M.; Hara, Y.; Kobayashi, S.; Iwase, H. Quantitation of HDAC1 mRNA Expression in Invasive Carcinoma of the Breast*. Breast Cancer Res. Treat. 2005, 94, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, A.; Horiuchi, A.; Kikuchi, N.; Hayashi, T.; Fuseya, C.; Suzuki, A.; Konishi, I.; Shiozawa, T. Type-specific roles of histone deacetylase (HDAC) overexpression in ovarian carcinoma: HDAC1 enhances cell proliferation and HDAC3 stimulates cell migration with downregulation of E-cadherin. Int. J. Cancer 2010, 127, 1332–1346. [Google Scholar] [CrossRef] [PubMed]
- Weichert, W.; Röske, A.; Gekeler, V.; Beckers, T.; Stephan, C.; Jung, K.; Fritzsche, F.R.; Niesporek, S.; Denkert, C.; Dietel, M.; et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br. J. Cancer 2008, 98, 604–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritzsche, F.R.; Weichert, W.; Röske, A.; Gekeler, V.; Beckers, T.; Stephan, C.; Jung, K.; Scholman, K.; Denkert, C.; Dietel, M.; et al. Class I histone deacetylases 1, 2 and 3 are highly expressed in renal cell cancer. BMC Cancer 2008, 8, 381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poyet, C.; Jentsch, B.; Hermanns, T.; Schweckendiek, D.; Seifert, H.-H.; Schmidtpeter, M.; Sulser, T.; Moch, H.; Wild, P.J.; Kristiansen, G. Expression of histone deacetylases 1, 2 and 3 in urothelial bladder cancer. BMC Clin. Pathol. 2014, 14, 10. [Google Scholar] [CrossRef] [Green Version]
- Moreno, D.A.; Scrideli, C.A.; Cortez, M.A.A.; Queiroz, R.D.P.; Valera, E.T.; Silveira, V.S.; Yunes, J.A.; Brandalise, S.R.; Tone, L.G. research paper: Differential expression of HDAC3, HDAC7 and HDAC9 is associated with prognosis and survival in childhood acute lymphoblastic leukaemia. Br. J. Haematol. 2010, 150, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C.; Kafeel, M.I.; Avezbakiyev, B.; Chen, C.; Sun, Y.; Rathnasabapathy, C.; Kalavar, M.; He, Z.; Burton, J.; Lichter, S. Histone deacetylase in chronic lymphocytic leukemia. Oncology 2012, 81, 325–329. [Google Scholar] [CrossRef]
- Ecker, J.; Oehme, I.; Mazitschek, R.; Korshunov, A.; Kool, M.; Hielscher, T.; Kiss, J.; Selt, F.; Konrad, C.; Lodrini, M.; et al. Targeting class I histone deacetylase 2 in MYC amplified group 3 medulloblastoma. Acta Neuropathol. Commun. 2015, 3, 22. [Google Scholar] [CrossRef] [Green Version]
- Ropero, S.; Fraga, M.F.; Ballestar, E.; Hamelin, R.; Yamamoto, H.; Boix-Chornet, M.; Caballero, R.; Alaminos, M.; Setien, F.; Paz, M.F.; et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat. Genet. 2006, 38, 566–569. [Google Scholar] [CrossRef]
- Müller, B.M.; Jana, L.; Kasajima, A.; Lehmann, A.; Prinzler, J.; Budczies, J.; Winzer, K.-J.; Dietel, M.; Weichert, W.; Denkert, C. Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer - overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression. BMC Cancer 2013, 13, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritsche, P.; Seidler, B.; Schüler, S.; Schnieke, A.; Göttlicher, M.; Schmid, R.M.; Saur, D.; Schneider, G. HDAC2 mediates therapeutic resistance of pancreatic cancer cells via the BH3-only protein NOXA. Gut 2009, 58, 1399–1409. [Google Scholar] [CrossRef] [Green Version]
- Minamiya, Y.; Ono, T.; Saito, H.; Takahashi, N.; Ito, M.; Motoyama, S.; Ogawa, J. Strong expression of HDAC3 correlates with a poor prognosis in patients with adenocarcinoma of the lung. Tumor Boil. 2010, 31, 533–539. [Google Scholar] [CrossRef]
- Wilmott, J.S.; Colebatch, A.J.; Kakavand, H.; Shang, P.; Carlino, M.S.; Thompson, J.; Long, G.V.; Scolyer, R.A.; Hersey, P. Expression of the class 1 histone deacetylases HDAC8 and 3 are associated with improved survival of patients with metastatic melanoma. Mod. Pathol. 2015, 28, 884–894. [Google Scholar] [CrossRef] [Green Version]
- Oehme, I.; Deubzer, H.E.; Wegener, D.; Pickert, D.; Linke, J.-P.; Hero, B.; Kopp-Schneider, A.; Westermann, F.; Ulrich, S.M.; Von Deimling, A.; et al. Histone Deacetylase 8 in Neuroblastoma Tumorigenesis. Clin. Cancer Res. 2009, 15, 91–99. [Google Scholar] [CrossRef] [Green Version]
- Song, C.; Zhu, S.; Wu, C.; Kang, J. Histone Deacetylase (HDAC) 10 Suppresses Cervical Cancer Metastasis through Inhibition of Matrix Metalloproteinase (MMP) 2 and 9 Expression*. J. Boil. Chem. 2013, 288, 28021–28033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minamiya, Y.; Ono, T.; Saito, H.; Takahashi, N.; Ito, M.; Mitsui, M.; Motoyama, S.; Ogawa, J. Expression of histone deacetylase 1 correlates with a poor prognosis in patients with adenocarcinoma of the lung. Lung Cancer 2011, 74, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Taunton, J.; Hassig, C.A.; Schreiber, S.L. A Mammalian Histone Deacetylase Related to the Yeast Transcriptional Regulator Rpd3p. Science 1996, 272, 408–411. [Google Scholar] [CrossRef]
- Yang, W.-M.; Inouye, C.; Zeng, Y.; Bearss, D.; Seto, E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. USA 1996, 93, 12845–12850. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.-M.; Yao, Y.-L.; Sun, J.-M.; Davie, J.R.; Seto, E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J. Boil. Chem. 1997, 272, 28001–28007. [Google Scholar] [CrossRef] [Green Version]
- Grozinger, C.M.; Hassig, C.A.; Schreiber, S.L. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl. Acad. Sci. USA 1999, 96, 4868–4873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kao, H.-Y.; Downes, M.; Ordentlich, P.; Evans, R.M. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genome Res. 2000, 14, 55–66. [Google Scholar]
- Hu, E.; Chen, Z.; Fredrickson, T.; Zhu, Y.; Kirkpatrick, R.; Zhang, G.-F.; Johanson, K.; Sung, C.-M.; Liu, R.; Winkler, J. Cloning and Characterization of a Novel Human Class I Histone Deacetylase That Functions as a Transcription Repressor. J. Boil. Chem. 2000, 275, 15254–15264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrie, K.; Guidez, F.; Howell, L.; Healy, L.; Waxman, S.; Greaves, M.; Zelent, A. The Histone Deacetylase 9 Gene Encodes Multiple Protein Isoforms. J. Boil. Chem. 2003, 278, 16059–16072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witt, O.; Deubzer, H.E.; Milde, T.; Oehme, I. HDAC family: What are the cancer relevant targets? Cancer Lett. 2009, 277, 8–21. [Google Scholar] [CrossRef]
- Kao, H.-Y.; Lee, C.-H.; Komarov, A.; Han, C.C.; Evans, R.M. Isolation and Characterization of Mammalian HDAC10, a Novel Histone Deacetylase. J. Boil. Chem. 2001, 277, 187–193. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Cueto, M.A.; Asselbergs, F.; Atadja, P. Cloning and Functional Characterization of HDAC11, a Novel Member of the Human Histone Deacetylase Family. J. Boil. Chem. 2002, 277, 25748–25755. [Google Scholar] [CrossRef] [Green Version]
- Marks, P.A. Histone deacetylase inhibitors: A chemical genetics approach to understanding cellular functions. Biochim. Biophys. Acta (BBA) Bioenerg. 2010, 1799, 717–725. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Myers, S.J.; Dingledine, R. Transcriptional repression by REST: Recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 1999, 2, 867–872. [Google Scholar] [CrossRef]
- Lagger, S.; Meunier, M.; Mikula, M.; Brunmeir, R.; Schlederer, M.; Artaker, M.; Pusch, O.; Egger, G.; Hagelkruys, A.; Mikulits, W.; et al. Crucial function of histone deacetylase 1 for differentiation of teratomas in mice and humans. EMBO J. 2010, 29, 3992–4007. [Google Scholar] [CrossRef] [Green Version]
- Macdonald, J.L.; Roskams, A.J. Histone deacetylases 1 and 2 are expressed at distinct stages of neuro-glial development. Dev. Dyn. 2008, 237, 2256–2267. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Cubizolles, F.; Zhang, Y.; Reichert, N.; Kohler, H.; Seiser, C.; Matthias, P. Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev. 2010, 24, 455–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leus, N.G.; Zwinderman, M.R.; Dekker, F.J. Histone deacetylase 3 (HDAC 3) as emerging drug target in NF-κB-mediated inflammation. Curr. Opin. Chem. Boil. 2016, 33, 160–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jänsch, N.; Meyners, C.; Muth, M.; Kopranovic, A.; Witt, O.; Oehme, I.; Meyer-Almes, F.-J. The enzyme activity of histone deacetylase 8 is modulated by a redox-switch. Redox Boil. 2018, 20, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Gregoretti, I.; Lee, Y.-M.; Goodson, H.V. Molecular Evolution of the Histone Deacetylase Family: Functional Implications of Phylogenetic Analysis. J. Mol. Boil. 2004, 338, 17–31. [Google Scholar] [CrossRef]
- Tsai, S.-C.; Seto, E. Regulation of Histone Deacetylase 2 by Protein Kinase CK2. J. Boil. Chem. 2002, 277, 31826–31833. [Google Scholar] [CrossRef] [Green Version]
- Jamaladdin, S.; Kelly, R.D.W.; O’Regan, L.; Dovey, O.M.; Hodson, G.E.; Millard, C.J.; Portolano, N.; Fry, A.M.; Schwabe, J.W.; Cowley, S. Histone deacetylase (HDAC) 1 and 2 are essential for accurate cell division and the pluripotency of embryonic stem cells. In Proceedings of the Proceedings of the National Academy of Sciences, Seattle, WA, USA, 3 June 2014; Volume 111, pp. 9840–9845. [Google Scholar]
- Dovey, O.M.; Foster, C.T.; Conte, N.; Edwards, S.A.; Edwards, J.M.; Singh, R.; Vassiliou, G.S.; Bradley, A.; Cowley, S.; Vasilliou, G. Histone deacetylase 1 and 2 are essential for normal T-cell development and genomic stability in mice. Blood 2013, 121, 1335–1344. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Ng, H.H.; Erdjument-Bromage, H.; Tempst, P.; Bird, A.; Reinberg, D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genome Res. 1999, 13, 1924–1935. [Google Scholar] [CrossRef]
- Kuo, M.H.; Allis, C.D. Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays 1998, 20, 615–626. [Google Scholar] [CrossRef]
- Burke, L.J.; Baniahmad, A. Co-repressors 2000. FASEB J. 2000, 14, 1876–1888. [Google Scholar] [CrossRef] [Green Version]
- Millard, C.J.; Watson, P.J.; Fairall, L.; Schwabe, J.W. Targeting Class I Histone Deacetylases in a “Complex” Environment. Trends Pharmacol. Sci. 2017, 38, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Bantscheff, M.; Hopf, C.; Savitski, M.M.; Dittmann, A.; Grandi, P.; Michon, A.-M.; Schlegl, J.; Abraham, Y.; Becher, I.; Bergamini, G.; et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat. Biotechnol. 2011, 29, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.D.W.; Chandru, A.; Watson, P.J.; Song, Y.; Blades, M.; Robertson, N.; Jamieson, A.G.; Schwabe, J.W.; Cowley, S. Histone deacetylase (HDAC) 1 and 2 complexes regulate both histone acetylation and crotonylation in vivo. Sci. Rep. 2018, 8, 14690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, R.D.; Cowley, S. The physiological roles of histone deacetylase (HDAC) 1 and 2: Complex co-stars with multiple leading parts. Biochem. Soc. Trans. 2013, 41, 741–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silverstein, R.A.; Ekwall, K. Sin3: A flexible regulator of global gene expression and genome stability. Curr. Genet. 2004, 47, 1–17. [Google Scholar] [CrossRef]
- Chaubal, A.; Pile, L.A. Same agent, different messages: Insight into transcriptional regulation by SIN3 isoforms. Epigenetics Chromatin 2018, 11, 17. [Google Scholar] [CrossRef]
- Grzenda, A.; Lomberk, G.; Zhang, J.-S.; Urrutia, R. Sin3: Master scaffold and transcriptional corepressor. Biochim. Biophys. Acta (BBA) Bioenerg. 2009, 1789, 443–450. [Google Scholar] [CrossRef] [Green Version]
- Smith, K.T.; Martin-Brown, S.A.; Florens, L.; Washburn, M.P.; Workman, J. Deacetylase Inhibitors Dissociate the Histone-Targeting ING2 Subunit from the Sin3 Complex. Chem. Boil. 2010, 17, 65–74. [Google Scholar] [CrossRef] [Green Version]
- Fleischer, T.C.; Yun, U.J.; Ayer, D. Identification and Characterization of Three New Components of the mSin3A Corepressor Complex. Mol. Cell. Boil. 2003, 23, 3456–3467. [Google Scholar] [CrossRef] [Green Version]
- Binda, O.; Roy, J.-S.; Branton, P.E. RBP1 Family Proteins Exhibit SUMOylation-Dependent Transcriptional Repression and Induce Cell Growth Inhibition Reminiscent of Senescence. Mol. Cell. Boil. 2006, 26, 1917–1931. [Google Scholar] [CrossRef] [Green Version]
- Nikolaev, A.Y.; Papanikolaou, N.A.; Li, M.; Qin, J.; Gu, W. Identification of a novel BRMS1-homologue protein p40 as a component of the mSin3A/p33ING1b/HDAC1 deacetylase complex. Biochem. Biophys. Res. Commun. 2004, 323, 1216–1222. [Google Scholar] [CrossRef] [PubMed]
- Shiio, Y.; Rose, D.W.; Aur, R.; Donohoe, S.; Aebersold, R.; Eisenman, R.N. Identification and Characterization of SAP25, a Novel Component of the mSin3 Corepressor Complex. Mol. Cell. Boil. 2006, 26, 1386–1397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nan, X.; Ng, H.H.; Johnson, C.A.; Laherty, C.D.; Turner, B.M.; Eisenman, R.N.; Bird, A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998, 393, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Brubaker, K.; Cowley, S.M.; Huang, K.; Loo, L.; Yochum, G.S.; Ayer, D.E.; Eisenman, R.N.; Radhakrishnan, I. Solution structure of the interacting domains of the Mad-Sin3 complex: Implications for recruitment of a chromatin-modifying complex. Cell 2000, 103, 655–665. [Google Scholar] [CrossRef] [Green Version]
- Spronk, A.C.; Tessari, M.; Kaan, A.M.; Jansen, J.F.; Vermeulen, M.; Stunnenberg, H.G.; Vuister, G.W. The Mad1-Sin3B interaction involves a novel helical fold. Nat. Genet. 2000, 7, 1100–1104. [Google Scholar] [CrossRef]
- Sif, S.; Saurin, A.; Imbalzano, A.N.; Kingston, R.E. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genome Res. 2001, 15, 603–618. [Google Scholar] [CrossRef] [Green Version]
- Deplus, R.; Delatte, B.; Schwinn, M.K.; Defrance, M.; Méndez, J.; Murphy, N.; Dawson, M.A.; Volkmar, M.; Putmans, P.; Calonne, E.; et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013, 32, 645–655. [Google Scholar] [CrossRef]
- Baltus, G.A.; Kowalski, M.P.; Tutter, A.V.; Kadam, S. A Positive Regulatory Role for the mSin3A-HDAC Complex in Pluripotency through Nanog and Sox2*. J. Boil. Chem. 2009, 284, 6998–7006. [Google Scholar] [CrossRef] [Green Version]
- Gambi, G.; Di Simone, E.; Basso, V.; Ricci, L.; Wang, R.; Verma, A.; Elemento, O.; Ponzoni, M.; Inghirami, G.; Icardi, L.; et al. The Transcriptional Regulator Sin3A Contributes to the Oncogenic Potential of STAT3. Cancer Res. 2019, 79, 3076–3087. [Google Scholar] [CrossRef] [Green Version]
- Dannenberg, J.-H.; David, G.; Zhong, S.; Van Der Torre, J.; Wong, W.H.; A DePinho, R. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genome Res. 2005, 19, 1581–1595. [Google Scholar] [CrossRef] [Green Version]
- Pile, L.A.; Spellman, P.T.; Katzenberger, R.J.; Wassarman, D.A. The SIN3 deacetylase complex represses genes encoding mitochondrial proteins: Implications for the regulation of energy metabolism. J. Biol. Chem. 2003, 278, 37840–37848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smits, A.H.; Jansen, P.W.T.C.; Poser, I.; Hyman, A.A.; Vermeulen, M. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 2012, 41, e28. [Google Scholar] [CrossRef]
- Torchy, M.; Hamiche, A.; Klaholz, B. Structure and function insights into the NuRD chromatin remodeling complex. Cell. Mol. Life Sci. 2015, 72, 2491–2507. [Google Scholar] [CrossRef]
- Millard, C.J.; Varma, N.; Saleh, A.; Morris, K.; Watson, P.J.; Bottrill, A.; Fairall, L.R.; Smith, C.J.; Schwabe, J.W. The structure of the core NuRD repression complex provides insights into its interaction with chromatin. eLife 2016, 5, 7285. [Google Scholar] [CrossRef] [PubMed]
- Schmidberger, J.W.; Sharifi Tabar, M.; Torrado, M.; Silva, A.P.; Landsberg, M.J.; Brillault, L.; AlQarni, S.; Zeng, Y.C.; Parker, B.L.; Low, J.K.; et al. The MTA1 subunit of the nucleosome remodeling and deacetylase complex can recruit two copies of RBBP4/7. Protein Sci. 2016, 25, 1472–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denslow, S.A.; Wade, P.A. The human Mi-2/NuRD complex and gene regulation. Oncogene 2007, 26, 5433–5438. [Google Scholar] [CrossRef] [Green Version]
- Tong, J.K.; Hassig, C.A.; Schnitzler, G.R.; Kingston, R.E.; Schreiber, S.L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 1998, 395, 917–921. [Google Scholar] [CrossRef]
- Le Guezennec, X.; Vermeulen, M.; Brinkman, A.B.; Hoeijmakers, W.A.M.; Cohen, A.; Lasonder, E.; Stunnenberg, H.G. MBD2/NuRD and MBD3/NuRD, Two Distinct Complexes with Different Biochemical and Functional Properties. Mol. Cell. Boil. 2006, 26, 843–851. [Google Scholar] [CrossRef] [Green Version]
- Saito, M.; Ishikawa, F. The mCpG-binding Domain of Human MBD3 Does Not Bind to mCpG but Interacts with NuRD/Mi2 Components HDAC1 and MTA2. J. Boil. Chem. 2002, 277, 35434–35439. [Google Scholar] [CrossRef] [Green Version]
- Hendrich, B.; Tweedie, S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003, 19, 269–277. [Google Scholar] [CrossRef]
- Aguilera, C.; Nakagawa, K.; Sancho, R.; Chakraborty, A.; Hendrich, B.; Behrens, A. C-Jun N-terminal phosphorylation antagonises recruitment of the Mbd3/NuRD repressor complex. Nature 2011, 469, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, R.L.; Tosti, L.; Radzisheuskaya, A.; Caballero, I.M.; Kaji, K.; Hendrich, B.; Silva, J.C. MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell 2014, 15, 102–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, E.; Han, C.; Park, I.; Lee, B.; Jin, S.; Choi, H.; Kim, H.; Park, Z.Y.; Eddy, E.M.; Cho, C. A Novel Germ Cell-specific Protein, SHIP1, Forms a Complex with Chromatin Remodeling Activity during Spermatogenesis*. J. Boil. Chem. 2008, 283, 35283–35294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujisaki, S.; Sato, A.; Toyomoto, T.; Hayano, T.; Sugai, M.; Kubota, T.; Koiwai, O. Direct binding of TReP-132 with TdT results in reduction of TdT activity. Genes Cells 2005, 11, 47–57. [Google Scholar] [CrossRef]
- You, A.; Tong, J.K.; Grozinger, C.M.; Schreiber, S.L. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc. Natl. Acad. Sci. USA 2001, 98, 1454–1458. [Google Scholar] [CrossRef] [Green Version]
- Fischle, W.; Dequiedt, F.; Hendzel, M.J.; Guenther, M.G.; Lazar, M.A.; Voelter, W.; Verdin, E. Enzymatic Activity Associated with Class II HDACs Is Dependent on a Multiprotein Complex Containing HDAC3 and SMRT/N-CoR. Mol. Cell 2002, 9, 45–57. [Google Scholar] [CrossRef] [Green Version]
- Lahm, A.; Paolini, C.; Pallaoro, M.; Nardi, M.C.; Jones, P.; Neddermann, P.; Sambucini, S.; Bottomley, M.J.; Surdo, P.L.; Carfí, A.; et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc. Natl. Acad. Sci. USA 2007, 104, 17335–17340. [Google Scholar] [CrossRef] [Green Version]
- Verdin, E.; Dequiedt, F.; Kasler, H.G. Class II histone deacetylases: Versatile regulators. Trends Genet. 2003, 19, 286–293. [Google Scholar] [CrossRef] [Green Version]
- Parra, M.; Verdin, E. Regulatory signal transduction pathways for class IIa histone deacetylases. Curr. Opin. Pharmacol. 2010, 10, 454–460. [Google Scholar] [CrossRef]
- Martin, M.; Kettmann, R.; Dequiedt, F. Class IIa histone deacetylases: Conducting development and differentiation. Int. J. Dev. Boil. 2009, 53, 291–301. [Google Scholar] [CrossRef]
- Martin, M.; Kettmann, R.; Dequiedt, F. Class IIa histone deacetylases: Regulating the regulators. Oncogene 2007, 26, 5450–5467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradner, J.E.; West, N.; Grachan, M.L.; Greenberg, E.F.; Haggarty, S.J.; Warnow, T.; Mazitschek, R. Chemical phylogenetics of histone deacetylases. Nat. Methods 2010, 6, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Guardiola, A.R.; Yao, T.-P. Molecular Cloning and Characterization of a Novel Histone Deacetylase HDAC10. J. Boil. Chem. 2001, 277, 3350–3356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotian, S.; Liyanarachchi, S.; Zelent, A.; Parvin, J.D. Histone Deacetylases 9 and 10 Are Required for Homologous Recombination*. J. Boil. Chem. 2011, 286, 7722–7726. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.J.; Seto, E. The Rpd3/Hda1 family of lysine deacetylases: From bacteria and yeast to mice and men. Nat. Rev. Mol. Cell. Biol. 2008, 9, 206–218. [Google Scholar] [CrossRef] [Green Version]
- Boyault, C.; Zhang, Y.; Fritah, S.; Caron, C.; Gilquin, B.; Kwon, S.H.; Garrido, C.; Yao, T.-P.; Vourc’H, C.; Matthias, P.; et al. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genome Res. 2007, 21, 2172–2181. [Google Scholar] [CrossRef] [Green Version]
- Valenzuela-Fernandez, A.; Cabrero, J.R.; Serrador, J.; Sánchez-Madrid, F. HDAC6: A key regulator of cytoskeleton, cell migration and cell–cell interactions. Trends Cell Boil. 2008, 18, 291–297. [Google Scholar] [CrossRef]
- Yang, W.-M.; Tsai, S.-C.; Wen, Y.-D.; Fejér, G.; Seto, E. Functional Domains of Histone Deacetylase-3. J. Boil. Chem. 2002, 277, 9447–9454. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.-J.; Seto, E. HATs and HDACs: From structure, function and regulation to novel strategies for therapy and prevention. Oncogene 2007, 26, 5310–5318. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [Green Version]
- Maison, C.; Bailly, D.; Peters, A.H.; Quivy, J.-P.; Roche, D.; Taddei, A.; Lachner, M.; Jenuwein, T.; Almouzni, G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002, 30, 329–334. [Google Scholar] [CrossRef]
- Barneda-Zahonero, B.; Parra, M. Histone deacetylases and cancer. Mol. Oncol. 2012, 6, 579–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Hu, Q.; Kaufman, A.; D’Ercole, A.J.; Ye, P. Developmental expression of histone deacetylase 11 in the murine brain. J. Neurosci. Res. 2008, 86, 537–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lian, Z.-R.; Xu, Y.-F.; Wang, X.-B.; Gong, J.; Liu, Z.-J. Suppression of Histone Deacetylase 11 Promotes Expression of IL-10 in Kupffer Cells and Induces Tolerance Following Orthotopic Liver Transplantation in Rats. J. Surg. Res. 2012, 174, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-J. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004, 32, 959–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheikh, B.N. Crafting the Brain—Role of Histone Acetyltransferases in Neural Development and Disease. Cell Tissue Res. 2014, 356, 553–573. [Google Scholar] [CrossRef]
- Gupta, A.; Guerin-Peyrou, T.G.; Sharma, G.G.; Park, C.; Agarwal, M.; Ganju, R.K.; Pandita, S.; Choi, K.; Sukumar, S.; Pandita, R.K.; et al. The Mammalian Ortholog of Drosophila MOF That Acetylates Histone H4 Lysine 16 Is Essential for Embryogenesis and Oncogenesis. Mol. Cell. Boil. 2007, 28, 397–409. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.F.; Fischle, W.; Verdin, E.; Greene, W.C. Duration of nuclear NF-κB action regulated by reversible acetylation. Science 2001, 293, 1653–1657. [Google Scholar] [CrossRef] [Green Version]
- Shivdasanl, R.A.; Mayer, E.L.; Orkin, S.H. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 1995, 373, 432–434. [Google Scholar] [CrossRef]
- Sebastiano, V.; Dalvai, M.; Gentile, L.; Schubart, K.; Sutter, J.; Wu, G.; Tapia, N.; Esch, D.; Ju, J.-Y.; Hübner, K.; et al. Oct1 regulates trophoblast development during early mouse embryogenesis. Development 2010, 137, 3551–3560. [Google Scholar] [CrossRef] [Green Version]
- Thomas, M.J.; Seto, E. Unlocking the mechanisms of transcription factor YY1: Are chromatin modifying enzymes the key? Gene 1999, 236, 197–208. [Google Scholar] [CrossRef]
- Lee, J.S.; Galvin, K.M.; See, R.H.; Eckner, R.; Livingston, D.; Moran, E.; Shi, Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995, 9, 1188–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malewicz, M.; Perlmann, T. Function of transcription factors at DNA lesions in DNA repair. Exp. Cell Res. 2014, 329, 94–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, A.J.; Oren, M. The first 30 years of p53: Growing ever more complex. Nat. Rev. Cancer 2009, 9, 749–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luna, R.M.D.O.; Wagner, D.S.; Lozano, G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995, 378, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-H.; Tsay, Y.-G.; Tan, B.C.; Lo, W.-Y.; Lee, S.-C. Identification and Characterization of a Novel p300-mediated p53 Acetylation Site, Lysine 305. J. Boil. Chem. 2003, 278, 25568–25576. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Li, M.; Tang, Y.; Laszkowska, M.; Roeder, R.G.; Gu, W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2004, 101, 2259–2264. [Google Scholar] [CrossRef] [Green Version]
- Appella, E.; Anderson, C.W. Post-translational modifications and activation of p53 by genotoxic stresses. JBIC J. Boil. Inorg. Chem. 2001, 268, 2764–2772. [Google Scholar] [CrossRef] [PubMed]
- Kastan, M.B.; Zhan, Q.; El-Deiry, W.; Carrier, F.; Jacks, T.; Walsh, W.V.; Plunkett, B.S.; Vogelstein, B.; Fornace, A.J. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 1992, 71, 587–597. [Google Scholar] [CrossRef]
- Smith, M.; Chen, I.; Zhan, Q.; Bae, I.; Chen, C.; Gilmer, T.; Kastan, M.; O’Connor, P.; Fornace, A.J. Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science 1994, 266, 1376–1380. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Su, F.; Chen, D.; Shiloh, A.; Gu, W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 2000, 408, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Kawaguchi, Y.; Lai, C.-H.; Kovacs, J.J.; Higashimoto, Y.; Appella, E.; Yao, T.-P. MDM2–HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002, 21, 6236–6245. [Google Scholar] [CrossRef]
- Beishline, K.; Kelly, C.M.; Olofsson, B.A.; Koduri, S.; Emrich, J.; Greenberg, R.A.; Azizkhan-Clifford, J. Sp1 Facilitates DNA Double-Strand Break Repair through a Nontranscriptional Mechanism. Mol. Cell. Boil. 2012, 32, 3790–3799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajput, P.; Pandey, V.; Kumar, V. Stimulation of ribosomal RNA gene promoter by transcription factor Sp1 involves active DNA demethylation by Gadd45-NER pathway. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2016, 1859, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, J.; Crofts, L.A.; Quinlan, K.G.; Czolij, R.; Perkins, A.; Crossley, M. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics 2006, 87, 474–482. [Google Scholar] [CrossRef] [Green Version]
- Suske, G. The Sp-family of transcription factors. Gene 1999, 238, 291–300. [Google Scholar] [CrossRef]
- Solomon, S.S.; Majumdar, G.; Martínez-Hernández, A.; Raghow, R. A critical role of Sp1 transcription factor in regulating gene expression in response to insulin and other hormones. Life Sci. 2008, 83, 305–312. [Google Scholar] [CrossRef]
- Li, L.; Davie, J.R. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann. Anat. Anat. Anz. 2010, 192, 275–283. [Google Scholar] [CrossRef]
- Li, G.; Xie, Q.; Yang, Z.; Wang, L.; Zhang, X.; Zuo, B.; Zhang, S.; Yang, A.; Jia, L. Sp1-mediated epigenetic dysregulation dictates HDAC inhibitor susceptibility of HER2-overexpressing breast cancer. Int. J. Cancer 2019, 145, 3285–3298. [Google Scholar] [CrossRef]
- Banerjee, A.; Mahata, B.; Dhir, A.; Mandal, T.K.; Biswas, K. Elevated histone H3 acetylation and loss of the Sp1–HDAC1 complex de-repress the GM2-synthase gene in renal cell carcinoma. J. Boil. Chem. 2018, 294, 1005–1018. [Google Scholar] [CrossRef] [Green Version]
- Juli, G.; Oliverio, M.; Bellizzi, D.; Cantafio, M.E.G.; Grillone, K.; Passarino, G.; Colica, C.; Nardi, M.; Rossi, M.; Procopio, A.; et al. Anti-tumor Activity and Epigenetic Impact of the Polyphenol Oleacein in Multiple Myeloma. Cancers 2019, 11, 990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Patel, V.K.; Jain, D.K.; Patel, P.; Rajak, H. Panobinostat as Pan-deacetylase Inhibitor for the Treatment of Pancreatic Cancer: Recent Progress and Future Prospects. Oncol. Ther. 2016, 4, 73–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hull, E.; Montgomery, M.; Leyva, K.J. HDAC Inhibitors as Epigenetic Regulators of the Immune System: Impacts on Cancer Therapy and Inflammatory Diseases. BioMed Res. Int. 2016, 2016, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falkenberg, K.J.; Johnstone, R.W. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 2014, 13, 673–691. [Google Scholar] [CrossRef] [PubMed]
- Ridinger, J.; Koeneke, E.; Kolbinger, F.R.; Koerholz, K.; Mahboobi, S.; Hellweg, L.; Gunkel, N.; Miller, A.K.; Peterziel, H.; Schmezer, P.; et al. Dual role of HDAC10 in lysosomal exocytosis and DNA repair promotes neuroblastoma chemoresistance. Sci. Rep. 2018, 8, 10039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilting, R.H.; Yanover, E.; Heideman, M.R.; Jacobs, H.; Horner, J.; Van Der Torre, J.; DePinho, R.A.; Dannenberg, J.-H. Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and haematopoiesis. EMBO J. 2010, 29, 2586–2597. [Google Scholar] [CrossRef] [Green Version]
- Santoro, F.; Botrugno, O.A.; Zuffo, R.D.; Pallavicini, I.; Matthews, G.M.; Cluse, L.; Barozzi, I.; Senese, S.; Fornasari, L.; Moretti, S.; et al. A dual role for Hdac1: Oncosuppressor in tumorigenesis, oncogene in tumor maintenance. Blood 2013, 121, 3459–3468. [Google Scholar] [CrossRef] [Green Version]
- Bolden, J.E.; Peart, M.J.; Johnstone, R.W. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006, 5, 769–784. [Google Scholar] [CrossRef]
- Li, Y.; Seto, E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb. Perspect. Med. 2016, 6, a026831. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, G.J.A.; Richmond, P.; Bunker, E.N.; Karman, S.S.; Azofeifa, J.; Garnett, A.T.; Xu, Q.; Wheeler, G.E.; Toomey, C.M.; Zhang, Q.; et al. Genome-wide dose-dependent inhibition of histone deacetylases studies reveal their roles in enhancer remodeling and suppression of oncogenic super-enhancers. Nucleic Acids Res. 2017, 46, 1756–1776. [Google Scholar] [CrossRef]
- Yasui, W.; Oue, N.; Ono, S.; Mitani, Y.; Ito, R.; Nakayama, H. Histone acetylation and gastrointestinal carcinogenesis. Ann. New York Acad. Sci. 2003, 983, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Seto, E. Deacetylation of Nonhistone Proteins by HDACs and the Implications in Cancer. Pharmacol. Ther. Cough 2011, 206, 39–56. [Google Scholar] [CrossRef]
- Banik, D.; Moufarrij, S.; Villagra, A. Immunoepigenetics Combination Therapies: An Overview of the Role of HDACs in Cancer Immunotherapy. Int. J. Mol. Sci. 2019, 20, 2241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, O.; Fotheringham, S.; Wood, V.; Stimson, L.; Zhang, C.; Pezzella, F.; Duvic, M.; Kerr, D.J.; La Thangue, N.B. HR23B is a biomarker for tumor sensitivity to HDAC inhibitor-based therapy. Proc. Natl. Acad. Sci. USA 2010, 107, 6532–6537. [Google Scholar] [CrossRef] [Green Version]
- Finnin, M.S.; Donigian, J.R.; Cohen, A.; Richon, V.M.; Rifkind, R.A.; Marks, P.A.; Breslow, R.; Pavletich, N.P. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 1999, 401, 188–193. [Google Scholar] [CrossRef]
- Mai, A.; Altucci, L. Epi-drugs to fight cancer: From chemistry to cancer treatment, the road ahead. Int. J. Biochem. Cell Boil. 2009, 41, 199–213. [Google Scholar] [CrossRef] [PubMed]
- Licciardi, P.V.; Ververis, K.; Hiong, A.; Karagiannis, T.C. Histone deacetylase inhibitors (HDACIs): Multitargeted anticancer agents. Boil. Targets Ther. 2013, 7, 47–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckschlager, T.; Plch, J.; Stiborová, M.; Hrabeta, J. Histone Deacetylase Inhibitors as Anticancer Drugs. Int. J. Mol. Sci. 2017, 18, 1414. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Eom, G.H. HDAC and HDAC Inhibitor: From Cancer to Cardiovascular Diseases. Chonnam Med. J. 2016, 52, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanneman, M.; Dranoff, G. Combining immunotherapy and targeted therapies in cancer treatment. Nat. Rev. Cancer 2012, 12, 237–251. [Google Scholar] [CrossRef] [Green Version]
- Montgomery, R.L.; Potthoff, M.J.; Haberland, M.; Qi, X.; Matsuzaki, S.; Humphries, K.M.; Richardson, J.A.; Bassel-Duby, R.; Olson, E. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J. Clin. Investig. 2008, 118, 3588–3597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.-H.; Hao, C.; Liu, P.; Tian, X.; Wang, L.-H.; Zhao, L.; Zhu, C.-M. Valproic acid inhibits tumor angiogenesis in mice transplanted with Kasumi-1 leukemia cells. Mol. Med. Rep. 2013, 9, 443–449. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Jiang, S.; Chen, J.; Su, S.B. Suberoylanilide hydroxamic acid suppresses inflammation-induced neovascularization. Can. J. Physiol. Pharmacol. 2014, 92, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Morales, J.; Magaña, M.J.R.; Carranza, D.; Ortiz-Ferrón, G.; Ruiz-Ruiz, C. HDAC inhibitors with different gene regulation activities depend on the mitochondrial pathway for the sensitization of leukemic T cells to TRAIL-induced apoptosis. Cancer Lett. 2010, 297, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Bolden, E.J.; Shi, W.; Jankowski, K.; Kan, C.-Y.; Cluse, L.; Martin, B.P.; MacKenzie, K.; Smyth, G.K.; Johnstone, R.W. HDAC inhibitors induce tumor-cell-selective pro-apoptotic transcriptional responses. Cell Death Dis. 2013, 4, e519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenny, R.G.; Ude, Z.; Docherty, J.R.; Marmion, C.J. Vorinostat and Belinostat, hydroxamate-based anti-cancer agents, are nitric oxide donors. J. Inorg. Biochem. 2020, 206, 110981. [Google Scholar] [CrossRef]
- Kroesen, M.; Gielen, P.R.; Brok, I.C.; Armandari, I.; Hoogerbrugge, P.M.; Adema, G.J. HDAC inhibitors and immunotherapy; a double edged sword? Oncotarget 2014, 5, 6558–6572. [Google Scholar] [CrossRef] [Green Version]
- Shukla, S.; Meeran, S.M.; Katiyar, S.K. Epigenetic regulation by selected dietary phytochemicals in cancer chemoprevention. Cancer Lett. 2014, 355, 9–17. [Google Scholar] [CrossRef] [Green Version]
- Sanaei, M.; Kavoosi, F.; Roustazadeh, A.; Shahsavani, H. In Vitro Effect of the Histone Deacetylase Inhibitor Valproic Acid on Viability and Apoptosis of the PLC/PRF5 Human Hepatocellular Carcinoma Cell Line. Asian Pac. J. Cancer Prev. 2018, 19, 2507–2510. [Google Scholar] [PubMed]
- Cortiguera, M.G.; García-Gaipo, L.; Wagner, S.D.; León, J.; Batlle-López, A.; Delgado, M.D. Suppression of BCL6 function by HDAC inhibitor mediated acetylation and chromatin modification enhances BET inhibitor effects in B-cell lymphoma cells. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Chao, M.-W.; Chang, L.-H.; Tu, H.-J.; Chang, C.-D.; Lai, M.-J.; Chen, Y.-Y.; Liou, J.-P.; Teng, C.-M.; Pan, S.-L. Combination treatment strategy for pancreatic cancer involving the novel HDAC inhibitor MPT0E028 with a MEK inhibitor beyond K-Ras status. Clin. Epigenetics 2019, 11, 85. [Google Scholar] [CrossRef] [PubMed]
- Bandolik, J.; Hamacher, A.; Schrenk, C.; Weishaupt, R.; Kassack, M.U. Class I-Histone Deacetylase (HDAC) Inhibition is Superior to pan-HDAC Inhibition in Modulating Cisplatin Potency in High Grade Serous Ovarian Cancer Cell Lines. Int. J. Mol. Sci. 2019, 20, 3052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Lyu, J.; Yang, E.J.; Liu, Y.; Wu, C.; Pardeshi, L.; Tan, K.; Chen, Q.; Xu, X.; Deng, C.-X.; et al. Class I histone deacetylase inhibition is synthetic lethal with BRCA1 deficiency in breast cancer cells. Acta Pharm. Sin. B 2019, 10, 615–627. [Google Scholar] [CrossRef]
- Groselj, B.; Ruan, J.-L.; Scott, H.; Gorrill, J.; Nicholson, J.; Kelly, J.; Anbalagan, S.; Thompson, J.; Stratford, M.R.; Jevons, S.J.; et al. Radiosensitization In Vivo by Histone Deacetylase Inhibition with No Increase in Early Normal Tissue Radiation Toxicity. Mol. Cancer Ther. 2017, 17, 381–392. [Google Scholar] [CrossRef] [Green Version]
- Paillas, S.; Then, C.K.; Kilgas, S.; Ruan, J.-L.; Thompson, J.; Elliott, A.; Smart, S.; Kiltie, A.E. The Histone Deacetylase Inhibitor Romidepsin Spares Normal Tissues While Acting as an Effective Radiosensitizer in Bladder Tumors in Vivo. Int. J. Radiat. Oncol. 2020, 107, 212–221. [Google Scholar] [CrossRef] [Green Version]
- Finkler, N.J.; Dizon, D.S.; Braly, P.; Micha, J.; Lassen, U.; Celano, P.; Glasspool, R.; Crowley, E.; Buhl-Jensen, P.; Penson, R.T. Phase II multicenter trial of the histone deactylase inhibitor (HDACi) belinostat, carboplatin and paclitaxel (BelCaP) in patients (pts) with relapsed epithelial ovarian cancer (EOC). J. Clin. Oncol. 2008, 26, 5519. [Google Scholar] [CrossRef]
- Gao, S.; Li, X.; Zang, J.; Xu, W.; Zhang, Y. Preclinical and Clinical Studies of Chidamide (CS055/HBI-8000), An Orally Available Subtype-selective HDAC Inhibitor for Cancer Therapy. Anti-Cancer Agents Med. Chem. 2017, 17, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Manero, G.; Yang, H.; Bueso-Ramos, C.; Ferrajoli, A.; Cortes, J.; Wierda, W.G.; Faderl, S.; Koller, C.; Morris, G.; Rosner, G.; et al. Phase 1 study of the histone deacetylase inhibitor vorinostat (suberoylanilide hydroxamic acid [SAHA]) in patients with advanced leukemias and myelodysplastic syndromes. Blood 2008, 111, 1060–1066. [Google Scholar] [CrossRef] [Green Version]
- Piekarz, R.; Frye, R.; Turner, M.; Wright, J.J.; Allen, S.; Kirschbaum, M.H.; Zain, J.; Prince, H.M.; Leonard, J.P.; Geskin, L.J.; et al. Phase II Multi-Institutional Trial of the Histone Deacetylase Inhibitor Romidepsin As Monotherapy for Patients With Cutaneous T-Cell Lymphoma. J. Clin. Oncol. 2009, 27, 5410–5417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niesvizky, R.; Ely, S.; Mark, T.; Aggarwal, S.; Gabrilove, J.L.; Wright, J.J.; Chen-Kiang, S.; Sparano, J.A. Phase 2 trial of the histone deacetylase inhibitor romidepsin for the treatment of refractory multiple myeloma. Cancer 2010, 117, 336–342. [Google Scholar] [CrossRef] [Green Version]
- San-Miguel, J.; Richardson, P.G.; Günther, A.; Sezer, O.; Siegel, D.; Bladé, J.; Leblanc, R.; Sutherland, H.; Sopala, M.; Mishra, K.K.; et al. Phase Ib Study of Panobinostat and Bortezomib in Relapsed or Relapsed and Refractory Multiple Myeloma. J. Clin. Oncol. 2013, 31, 3696–3703. [Google Scholar] [CrossRef] [PubMed]
- Yazbeck, V.; Shafer, D.; Perkins, E.B.; Coppola, M.; Sokol, L.; Richards, K.L.; Shea, T.; Ruan, J.; Parekh, S.; Strair, R.; et al. A Phase II Trial of Bortezomib and Vorinostat in Mantle Cell Lymphoma and Diffuse Large B-cell Lymphoma. Clin. Lymphoma Myeloma Leuk. 2018, 18, 569–575.e1. [Google Scholar] [CrossRef] [PubMed]
- Houtkooper, R.H.; Pirinen, E.; Auwerx, J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Boil. 2012, 13, 225–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guha, M. HDAC inhibitors still need a home run, despite recent approval. Nat. Rev. Drug Discov. 2015, 14, 225–226. [Google Scholar] [CrossRef] [PubMed]
- Suraweera, A.; O’Byrne, K.J.; Richard, D.J. Combination Therapy With Histone Deacetylase Inhibitors (HDACi) for the Treatment of Cancer: Achieving the Full Therapeutic Potential of HDACi. Front. Oncol. 2018, 8, 92. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.H. Health-Promoting Components of Fruits and Vegetables in the Diet12. Adv. Nutr. 2013, 4, 384S–392S. [Google Scholar] [CrossRef]
- Thomas, T.R.A.; Kavlekar, D.P.; LokaBharathi, P.A. Marine Drugs from Sponge-Microbe Association—A Review. Mar. Drugs 2010, 8, 1417–1468. [Google Scholar] [CrossRef] [Green Version]
- Piña, I.C.; Gautschi , J.T.; Wang, G.Y.; Sanders, M.L.; Schmitz, F.J.; France, D.; Cornell-Kennon, S.; Sambucetti, L.C.; Remiszewski, S.W.; Perez, L.B.; et al. Psammaplins from the Sponge Pseudoceratina p urpurea: Inhibition of Both Histone Deacetylase and DNA Methyltransferase. J. Org. Chem. 2007, 68, 3866–3873. [Google Scholar]
- Taori, K.; Paul, V.J.; Luesch, H. Structure and activity of largazole, a potent antiproliferative agent from the Floridian marine cyanobacterium Symploca sp. J. Am. Chem. Soc. 2008, 130, 1806–1807. [Google Scholar] [CrossRef]
- Ying, Y.; Taori, K.; Kim, H.; Hong, J.; Luesch, H. Total synthesis and molecular target of largazole, a histone deacetylase inhibitor. J. Am. Chem. Soc. 2008, 130, 8455–8459. [Google Scholar] [CrossRef]
- Ueda, H.; Nakajima, H.; Hori, Y.; Fujita, T.; Nishimura, M.; Goto, T.; Okuhara, M. FR901228, a novel antitumor bicyclic depsipeptide produced by chromobacterium violaceum No. 968. I. Taxonomy, fermentation, isolation, physico-chemical and biological properties, and antitumor activity. J. Antibiot (Tokyo) 2012, 47, 301–310. [Google Scholar]
- Syed, D.N.; Adhami, V.M.; Khan, N.; Khan, M.I.; Mukhtar, H. Exploring the molecular targets of dietary flavonoid fisetin in cancer. Semin. Cancer Boil. 2016, 40–41, 130–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinha, D.; Sarkar, N.; Biswas, J.; Bishayee, A. Resveratrol for breast cancer prevention and therapy: Preclinical evidence and molecular mechanisms. Semin. Cancer Boil. 2016, 209–232. [Google Scholar] [CrossRef] [PubMed]
- Duvoix, A.; Blasius, R.; Delhalle, S.; Schnekenburger, M.; Morceau, F.; Henry, E.; Dicato, M.; Diederich, M. Chemopreventive and therapeutic effects of curcumin. Cancer Lett. 2005, 223, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Menezes, J.C.J.M.D.S.; Orlíková, B.; Morceau, F.; Diederich, M. Natural and Synthetic Flavonoids: Structure–Activity Relationship and Chemotherapeutic Potential for the Treatment of Leukemia. Crit. Rev. Food Sci. Nutr. 2015, 56, S4–S28. [Google Scholar] [CrossRef]
- Ko, J.-H.; Sethi, G.; Um, J.-Y.; Shanmugam, M.K.; Arfuso, F.; Kumar, A.P.; Bishayee, A.; Ahn, K.S. The Role of Resveratrol in Cancer Therapy. Int. J. Mol. Sci. 2017, 18, 2589. [Google Scholar] [CrossRef] [Green Version]
- Baek, S.-H.; Ko, J.-H.; Lee, H.; Jung, J.; Kong, M.; Lee, J.-W.; Lee, J.; Chinnathambi, A.; Zayed, M.; Alharbi, S.A.; et al. Resveratrol inhibits STAT3 signaling pathway through the induction of SOCS-1: Role in apoptosis induction and radiosensitization in head and neck tumor cells. Phytomedicine 2016, 23, 566–577. [Google Scholar] [CrossRef]
- Harikumar, K.B.; Kunnumakkara, A.B.; Sethi, G.; Diagaradjane, P.; Anand, P.; Pandey, M.K.; Gelovani, J.; Krishnan, S.; Guha, S.; Aggarwal, B.B. Resveratrol, a multitargeted agent, can enhance antitumor activity of gemcitabinein vitroand in orthotopic mouse model of human pancreatic cancer. Int. J. Cancer 2010, 127, 257–268. [Google Scholar] [CrossRef] [Green Version]
- Dhar, S.; Kumar, A.; Li, K.; Tzivion, G.; Levenson, A.S. Resveratrol regulates PTEN/Akt pathway through inhibition of MTA1/HDAC unit of the NuRD complex in prostate cancer. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 265–275. [Google Scholar] [CrossRef] [Green Version]
- Venturelli, S.; Berger, A.; Böcker, A.; Busch, C.; Weiland, T.; Noor, S.; Leischner, C.; Schleicher, S.; Mayer, M.; Weiss, T.S.; et al. Resveratrol as a Pan-HDAC Inhibitor Alters the Acetylation Status of Jistone Proteins in Human-Derived Hepatoblastoma Cells. PLoS ONE 2013, 8, e73097. [Google Scholar] [CrossRef]
- Bose, P.; Dai, Y.; Grant, S. Histone deacetylase inhibitor (HDACI) mechanisms of action: Emerging insights. Pharmacol. Ther. 2014, 143, 323–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bots, M.; Verbrugge, I.; Martin, B.P.; Salmon, J.M.; Ghisi, M.; Baker, A.; Stanley, K.; Shortt, J.; Ossenkoppele, G.J.; Zuber, J.; et al. Differentiation therapy for the treatment of t(8;21) acute myeloid leukemia using histone deacetylase inhibitors. Blood 2014, 123, 1341–1352. [Google Scholar] [CrossRef] [PubMed]
- Helland, Ø.; Popa, M.; Bischof, K.; Gjertsen, B.T.; Mc Cormack, E.; Bjørge, L. The HDACi Panobinostat Shows Growth Inhibition Both In Vitro and in a Bioluminescent Orthotopic Surgical Xenograft Model of Ovarian Cancer. PLOS ONE 2016, 11, e0158208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, L.; Sinha, R.; Itokawa, H.; Bastow, K.F.; Tachibana, Y.; Nakanishi, Y.; Kilgore, N.; Lee, K.H. Anti-HIV and cytotoxic activities of Ru(II)/Ru(III) polypyridyl complexes containing 2,6-(2′-benzimidazolyl)-pyridine/chalcone as co-ligand. Bioorganic Med. Chem. 2001, 9, 1667–1671. [Google Scholar] [CrossRef]
- Zhang, E.-H.; Wang, R.-F.; Guo, S.; Liu, B. An Update on Antitumor Activity of Naturally Occurring Chalcones. Evidence-Based Complement. Altern. Med. 2013, 2013, 1–22. [Google Scholar] [CrossRef]
- Jeon, J.-H.; Kim, S.-J.; Kim, C.-G.; Kim, J.-K.; Jun, J.-G. Synthesis of Biologically Active Chalcones and their Anti-inflammatory Effects. Bull. Korean Chem. Soc. 2012, 33, 953–957. [Google Scholar] [CrossRef] [Green Version]
- Kantevari, S.; Addla, D.; Bagul, P.K.; Sridhar, B.; Banerjee, S.K. Synthesis and evaluation of novel 2-butyl-4-chloro-1-methylimidazole embedded chalcones and pyrazoles as angiotensin converting enzyme (ACE) inhibitors. Bioorganic Med. Chem. 2011, 19, 4772–4781. [Google Scholar] [CrossRef]
- Detsi, A.; Majdalani, M.; Kontogiorgis, C.A.; Hadjipavlou-Litina, D.; Kefalas, P. Natural and synthetic 2′-hydroxy-chalcones and aurones: Synthesis, characterization and evaluation of the antioxidant and soybean lipoxygenase inhibitory activity. Bioorganic Med. Chem. 2009, 17, 8073–8085. [Google Scholar] [CrossRef] [PubMed]
- Romagnolo, D.F.; Selmin, O.I. Flavonoids and Cancer Prevention: A Review of the Evidence. J. Nutr. Gerontol. Geriatr. 2012, 31, 206–238. [Google Scholar] [CrossRef]
- Israf, D.; Khaizurin, T.; Syahida, A.; Lajis, N.; Khozirah, S. Cardamonin inhibits COX and iNOS expression via inhibition of p65NF-κB nuclear translocation and Iκ-B phosphorylation in RAW 264.7 macrophage cells. Mol. Immunol. 2007, 44, 673–679. [Google Scholar] [CrossRef]
- Mahapatra, D.K.; Asati, V.; Bharti, S.K. ChemInform Abstract: Chalcones and Their Therapeutic Targets for the Management of Diabetes: Structural and Pharmacological Perspectives. Cheminform 2015, 46, 839–865. [Google Scholar] [CrossRef]
- Syam, S.; Abdelwahab, S.; Al-Mamary, M.A.; Mohan, S. Synthesis of Chalcones with Anticancer Activities. Molecules 2012, 17, 6179–6195. [Google Scholar] [CrossRef]
- Pande, A.N.; Biswas, S.; Reddy, N.D.; Jayashree, B.S.; Kumar, N.; Rao, C.M. In vitro and in vivo anticancer studies of 2′-hydroxy chalcone derivatives exhibit apoptosis in colon cancer cells by HDAC inhibition and cell cycle arrest. EXCLI J. 2017, 16, 448. [Google Scholar]
- Zhou, J.; Li, M.; Chen, N.; Wang, S.; Luo, H.-B.; Zhang, Y.; Wu, R. Computational Design of a Time-Dependent Histone Deacetylase 2 Selective Inhibitor. ACS Chem. Boil. 2015, 10, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Orlikova, B.; Schnekenburger, M.; Zloh, M.; Golais, F.; Diederich, M.; Tasdemir, D. Natural chalcones as dual inhibitors of HDACs and NF-κB. Oncol. Rep. 2012, 28, 797–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahyo, T.; Ichikawa, S.; Hatanaka, T.; Yamada, M.; Setou, M. A novel chalcone polyphenol inhibits the deacetylase activity of SIRT1 and cell growth in HEK293T cells. J. Pharmacol. Sci. 2008, 108, 364–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, M.F.; Shaykoon, M.S.; Abdelrahman, M.H.; Elsadek, B.E.M.; Aboraia, A.S.; Abuo-Rahma, G.E.-D.A. Design, synthesis, docking studies and biological evaluation of novel chalcone derivatives as potential histone deacetylase inhibitors. Bioorganic Chem. 2017, 72, 32–41. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Sung, B. Pharmacological basis for the role of curcumin in chronic diseases: An age-old spice with modern targets. Trends Pharmacol. Sci. 2009, 30, 85–94. [Google Scholar] [CrossRef]
- Shehzad, A.; Shahzad, R.; Lee, Y.S. Curcumin: A Potent Modulator of Multiple Enzymes in Multiple Cancers. Enzymes 2014, 36, 149–174. [Google Scholar]
- Sung, B.; Kunnumakkara, A.B.; Sethi, G.; Anand, P.; Guha, S.; Aggarwal, B.B. Curcumin circumvents chemoresistance in vitro and potentiates the effect of thalidomide and bortezomib against human multiple myeloma in nude mice model. Mol. Cancer Ther. 2009, 8, 959–970. [Google Scholar] [CrossRef] [Green Version]
- Kamat, A.M.; Sethi, G.; Aggarwal, B.B. Curcumin potentiates the apoptotic effects of chemotherapeutic agents and cytokines through down-regulation of nuclear factor- B and nuclear factor- B-regulated gene products in IFN- -sensitive and IFN- -resistant human bladder cancer cells. Mol. Cancer Ther. 2007, 6, 1022–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shishodia, S.; Sethi, G.; Aggarwal, B.B. Curcumin: Getting Back to the Roots. Ann. New York Acad. Sci. 2005, 1056, 206–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandur, S.K.; Pandey, M.K.; Sung, B.; Ahn, K.S.; Murakami, A.; Sethi, G.; Limtrakul, P.; Badmaev, V.; Aggarwal, B.B. Curcumin, demethoxycurcumin, bisdemethoxycurcumin, tetrahydrocurcumin and turmerones differentially regulate anti-inflammatory and anti-proliferative responses through a ROS-independent mechanism. Carcinogenesis 2007, 28, 1765–1773. [Google Scholar] [CrossRef] [PubMed]
- Lao, C.D.; Ruffin, M.; Normolle, D.P.; Heath, D.D.; Murray, S.I.; Bailey, J.M.; Boggs, M.E.; Crowell, J.; Rock, C.L.; Brenner, D.E. Dose escalation of a curcuminoid formulation. BMC Complement. Altern. Med. 2006, 6, 10. [Google Scholar] [CrossRef] [Green Version]
- Hatcher, H.; Planalp, R.; Cho, J.; Torti, F.M.; Torti, S.V. Curcumin: From ancient medicine to current clinical trials. Cell. Mol. Life Sci. 2008, 65, 1631–1652. [Google Scholar] [CrossRef] [PubMed]
- Surh, Y.-J.; Chun, K.-S.; Cha, H.-H.; Han, S.S.; Keum, Y.-S.; Park, K.-K.; Lee, S.S. Molecular mechanisms underlying chemopreventive activities of anti-inflammatory phytochemicals: Down-regulation of COX-2 and iNOS through suppression of NF-κB activation. Mutat. Res. Mol. Mech. Mutagen. 2001, 480, 243–268. [Google Scholar] [CrossRef]
- Skommer, J.; Wlodkowic, D.; Pelkonen, J. Gene-expression profiling during curcumin-induced apoptosis reveals downregulation of CXCR4. Exp. Hematol. 2007, 35, 84–95. [Google Scholar] [CrossRef]
- Teiten, M.-H.; Dicato, M.; Diederich, M. Curcumin as a regulator of epigenetic events. Mol. Nutr. Food Res. 2013, 57, 1619–1629. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef]
- Wang, S.-H.; Lin, P.-Y.; Chiu, Y.-C.; Huang, J.-S.; Kuo, Y.-T.; Wu, J.-C.; Chen, C.-C. Curcumin-Mediated HDAC Inhibition Suppresses the DNA Damage Response and Contributes to Increased DNA Damage Sensitivity. PLOS ONE 2015, 10, e0134110. [Google Scholar] [CrossRef]
- Zeng, L.-L.; Shu, W.; Chen, W.; Wu, Q.; Liu, H.; Cui, G. Curcumin, both Histone Deacetylase and p300/CBP-Specific Inhibitor, Represses the Activity of Nuclear Factor Kappa B and Notch 1 in Raji Cells. Basic Clin. Pharmacol. Toxicol. 2007, 101, 427–433. [Google Scholar] [CrossRef]
- Liu, H.-L.; Chen, Y.; Cui, G.-H.; Zhou, J.-F. Curcumin, a potent anti-tumor reagent, is a novel histone deacetylase inhibitor regulating B-NHL cell line Raji proliferation. Acta Pharmacol. Sin. 2005, 26, 603–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.Q.; Yu, K.; Yan, Q.X.; Xing, C.Y.; Chen, Y.; Yan, Z.; Shi, Y.F.; Zhao, K.W.; Gao, S.M. Pure curcumin increases the expression of SOCS1 and SOCS3 in myeloproliferative neoplasms through suppressing class I histone deacetylases. Carcinogenesis 2013, 34, 1442–1449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liou, W.-S.; Lin, C.; Lee, P.-S.; Kalyanam, N.; Ho, C.-T.; Pan, M.-H. Calebin-A induces cell cycle arrest in human colon cancer cells and xenografts in nude mice. J. Funct. Foods 2016, 26, 781–791. [Google Scholar] [CrossRef]
- Oliveira, A.L.D.P.; Martinez, S.E.; Nagabushnam, K.; Majeed, M.; Alrushaid, S.; Sayre, C.; Davies, N.M.; Oliveir, A.L.D.P.; Martine, S.E.; Nagabushna, K.; et al. Calebin A: Analytical Development for Pharmacokinetics Study, Elucidation of Pharmacological Activities and Content Analysis of Natural Health Products. J. Pharm. Pharm. Sci. 2015, 18, 494. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.S.; Parmigiani, R.B.; Marks, P.A. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene 2007, 26, 5541–5552. [Google Scholar] [CrossRef] [Green Version]
- Qing, W.; Yan, C.; Xinggang, L. HDAC1 expression and effect of curcumin on proliferation of Raji cells. Acta Acad. Med. Wuhan 2006, 26, 199–201. [Google Scholar] [CrossRef]
- Sarkar, R.; Mukherjee, A.; Mukherjee, S.; Biswas, R.; Biswas, J.; Roy, M. Curcumin augments the efficacy of antitumor drugs used in leukemia by modulation of heat shock proteins via HDAC6. J. Environ. Pathol. Toxicol. Oncol. 2014, 33, 247–263. [Google Scholar] [CrossRef]
- Bhullar, K.S.; Jha, A.; Rupasinghe, H.V. Novel carbocyclic curcumin analog CUR3d modulates genes involved in multiple apoptosis pathways in human hepatocellular carcinoma cells. Chem. Interactions 2015, 242, 107–122. [Google Scholar] [CrossRef]
- Shu, L.; Khor, T.O.; Lee, J.H.; Boyanapalli, S.S.S.; Huang, Y.; Wu, T.-Y.; Saw, C.; Cheung, K.-L.; Kong, A.-N. Epigenetic CpG Demethylation of the Promoter and Reactivation of the Expression of Neurog1 by Curcumin in Prostate LNCaP Cells. AAPS J. 2011, 13, 606–614. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Shu, L.; Zhang, C.; Su, Z.-Y.; Kong, A.-N. Curcumin inhibits anchorage-independent growth of HT29 human colon cancer cells by targeting epigenetic restoration of the tumor suppressor gene DLEC1. Biochem. Pharmacol. 2015, 94, 69–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soflaei, S.S.; Momtazi-Borojeni, A.A.; Majeed, M.; DeRosa, G.; Maffioli, P.; Sahebkar, A. Curcumin: A Natural Pan-HDAC Inhibitor in Cancer. Curr. Pharm. Desn 2018, 24, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.R.; Sangeetha, S.; Ranjitha, S.; Murugan, K. Breast Cancer Specific Histone Deacetylase Inhibitors and Lead Discovery using Molecular Docking and Descriptor Study. Trends Bioinform. 2013, 6, 25–44. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.-J.; Tsai, Y.-J.; Lin, M.-Y.; You, H.-L.; Kalyanam, N.; Ho, C.-T.; Pan, M.-H. Calebin-A induced death of malignant peripheral nerve sheath tumor cells by activation of histone acetyltransferase. Phytomedicine 2019, 57, 377–384. [Google Scholar] [CrossRef]
- Novaes, J.T.; Lillico, R.; Sayre, C.; Nagabhushanam, K.; Majeed, M.; Chen, Y.; Ho, E.; Oliveira, A.L.D.P.; Martinez, S.E.; Alrushaid, S.; et al. Disposition, Metabolism and Histone Deacetylase and Acetyltransferase Inhibition Activity of Tetrahydrocurcumin and Other Curcuminoids. Pharmaceutics 2017, 9, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pari, L.; Amali, D.R. Protective role of tetrahydrocurcumin (THC) an active principle of turmeric on chloroquine induced hepatotoxicity in rats. J. Pharm. Pharm. Sci. 2005, 8, 115–123. [Google Scholar] [PubMed]
- Aggarwal, B.B.; Deb, L.; Prasad, S. Curcumin Differs from Tetrahydrocurcumin for Molecular Targets, Signaling Pathways and Cellular Responses. Molecules 2014, 20, 185–205. [Google Scholar] [CrossRef] [Green Version]
- Bora-Tatar, G.; Dayangac-Erden, D.; Demir, A.S.; Dalkara, S.; Yelekçi, K.; Yurter, H.E. Molecular modifications on carboxylic acid derivatives as potent histone deacetylase inhibitors: Activity and docking studies. Bioorganic Med. Chem. 2009, 17, 5219–5228. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Verner, E.; Buggy, J.J. Isoform-specific histone deacetylase inhibitors: The next step? Cancer Lett. 2009, 280, 211–221. [Google Scholar] [CrossRef]

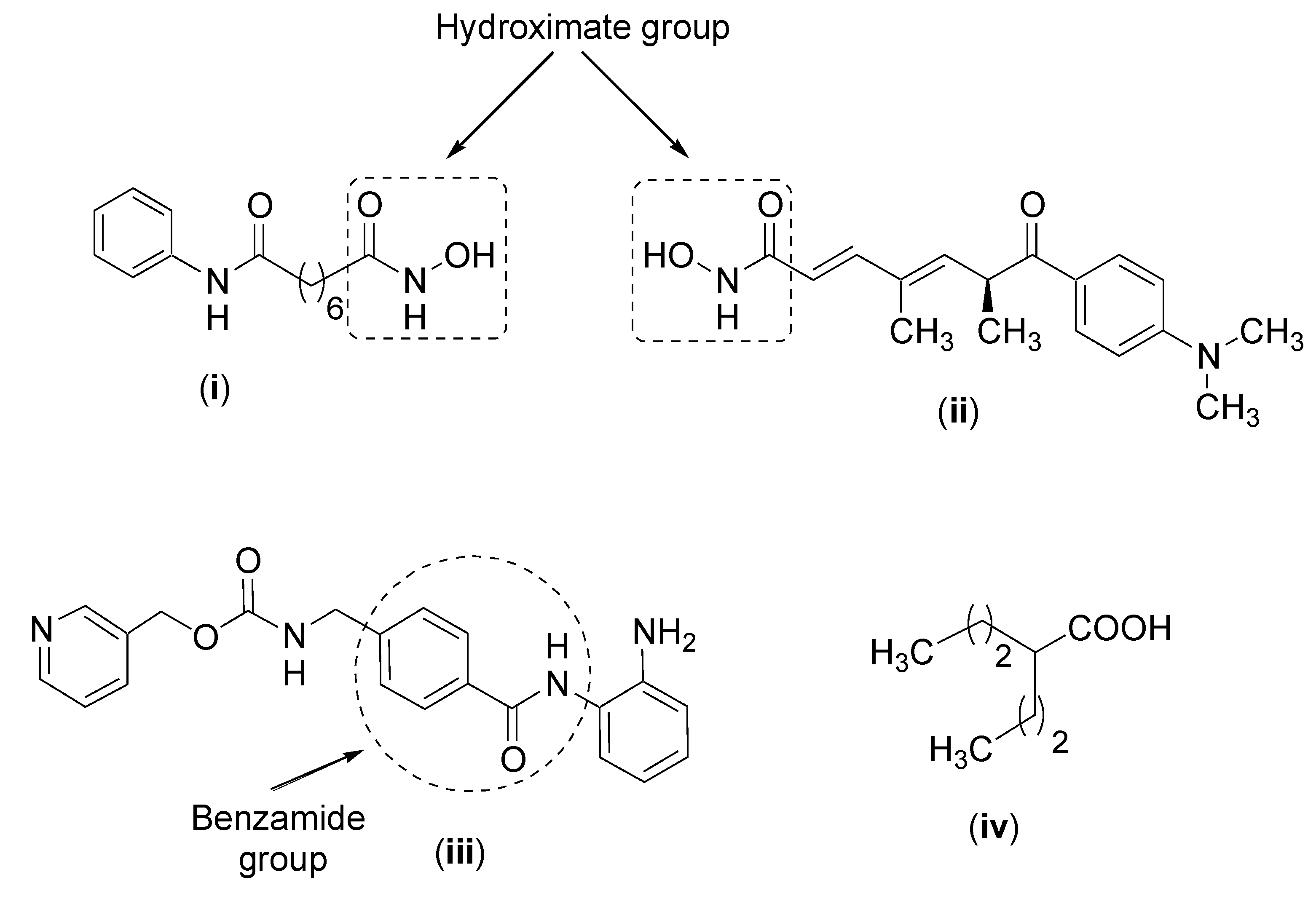
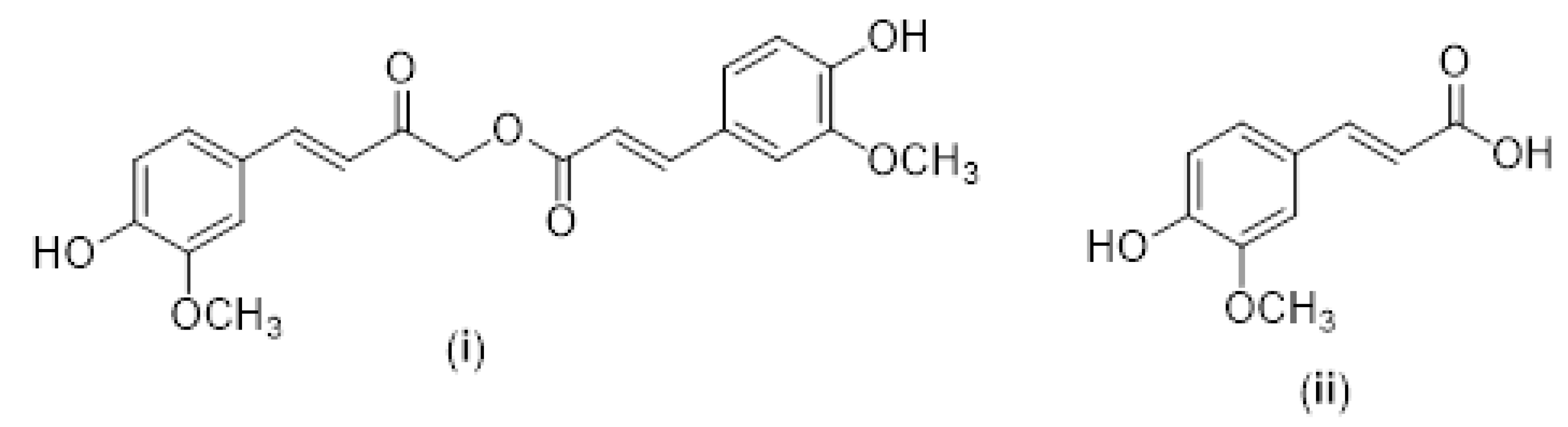
| HDAC Type | HDAC Class | Amino Acids | Cellular Localization | Cancer Type | Referencers |
|---|---|---|---|---|---|
| HDAC1 | I | 482 | Nucleus | Lung, gastric, liver, breast, ovarian, prostate, renal, bladder, hematological tumors | [52,53,54,55,56,57,58,59] |
| HDAC2 | I | 488 | Nucleus | Medulloblastoma, gastric, pancreatic, colorectal, breast, ovarian, prostate, renal, bladder, hematological tumors | [54,55,56,57,58,60,61,62,63] |
| HDAC3 | I | 428 | Nucleus and cytoplasm | Lung, gastric, breast, ovarian, prostate, bladder, melanoma, Hematological tumors | [54,55,57,58,62,64,65] |
| HDAC4 | IIa | 1084 | Nucleus and cytoplasm | Hematological tumors | [58] |
| HDAC5 | IIa | 1122 | Nucleus and cytoplasm | Hematological tumors | [58] |
| HDAC6 | IIb | 1215 | cytoplasm | Hematological tumors | [58] |
| HDAC7 | IIa | 912 | Nucleus and cytoplasm | Hematological tumors | [58] |
| HDAC8 | I | 377 | Nucleus, Mitochondria and cytoplasm | Neuroblastoma, melanoma, hematological tumors | [58,65,66] |
| HDAC9 | IIa | 1011 | Nucleus and cytoplasm | Hematological tumors | [58] |
| HDAC10 | IIb | 669 | Cytoplasm | Cervical, chronic lymphocytic leukemia | [59,67] |
| HDAC11 | IV | 347 | Nucleus | Lung | [68] |
| Cell Line | Curcumin | Molecular Effects | Result | References |
|---|---|---|---|---|
| Lymphoma Raji cells | 1.6–50 μM | Cell proliferation | ↓HDAC1 mRNA and protein expression | [269] |
| Human cervical cancer cell lines | 0.5–50 μM | Augments the efficacy of antitumor drugs | ↓HDAC activity, ↓HDAC1and 2 expression | [270] |
| HepG2 cell line | 100 μM of different curcumin analogues | Modulates genes | ↓HDAC1, 2, 4, 6, 8, 11 expression | [271] |
| K562, HEL, and MPN cell lines | 5–40 μM | Increases the expression of suppressor of cytokine signaling 1 and 3 | ↓HDAC activity ↓HDAC1, 3, 8 expression | [265] |
| LNCaP cells | 5 μM | CpG demethylation | ↓HDAC activity ↑HDAC1, 4, 5 and 8 and ↓HDAC3 protein expression | [272] |
| HT29 cell line | 2.5 and 5 μM | Inhibitory effect on cell proliferation | ↓HDAC4, 5, 6 and 8 expression | [273] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verza, F.A.; Das, U.; Fachin, A.L.; Dimmock, J.R.; Marins, M. Roles of Histone Deacetylases and Inhibitors in Anticancer Therapy. Cancers 2020, 12, 1664. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12061664
Verza FA, Das U, Fachin AL, Dimmock JR, Marins M. Roles of Histone Deacetylases and Inhibitors in Anticancer Therapy. Cancers. 2020; 12(6):1664. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12061664
Chicago/Turabian StyleVerza, Flávia Alves, Umashankar Das, Ana Lúcia Fachin, Jonathan R. Dimmock, and Mozart Marins. 2020. "Roles of Histone Deacetylases and Inhibitors in Anticancer Therapy" Cancers 12, no. 6: 1664. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12061664






