Systematic Review of Pharmacogenetic Factors That Influence High-Dose Methotrexate Pharmacokinetics in Pediatric Malignancies
Abstract
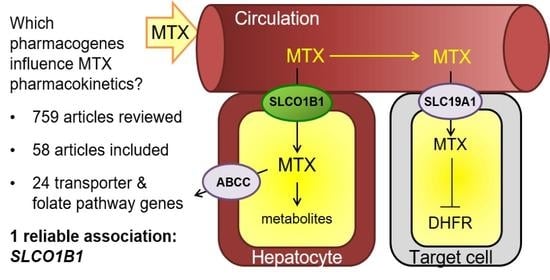
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Eligibility Criteria
- Evaluated the association between genetic polymorphisms and MTX PK;
- Included pediatric patients
- With ALL, lymphoma, or osteosarcoma.
- Only investigated the PK of MTX (no genetics),
- Only investigated the pharmacogenetics of MTX on toxicity (no PK),
- Only investigated patients with other malignancies,
- Did not include pediatric patients (only include adult populations), and
- Were not published in English.
2.2. Search Strategy
2.3. Data Collection
2.4. Qualitative Data Analysis
3. Results
3.1. Study Selection
3.2. Study Characteristics
3.3. Association between Genetic Polymorphisms and MTX PKs
3.3.1. Transporter Genes
ABCB1
ABCG2
ABCC2
ABCC3
ABCC4
ABCC1, ABCC5 and ABCC10
SLC19A1
SLC22A6 and SLCO22A8
SLCO1A2
SLCO1B1
SLCO1B3
3.3.2. Folate Pathway Genes
ATIC
ARID5B
DHFR
GGH
FPGS
MTHFD1
MTHFR
MTR
MTRR
TYMS
3.3.3. Additional Genes Examined
4. Discussion
4.1. Toxicity
4.2. Limitations and Future Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jolivet, J.; Cowan, K.H.; Curt, G.A.; Clendeninn, N.J.; Chabner, B.A. The pharmacology and clinical use of methotrexate. N. Engl. J. Med. 1983, 309, 1094–1104. [Google Scholar] [CrossRef]
- Evans, W.E.; Crom, W.R.; Abromowitch, M.; Dodge, R.; Look, A.T.; Bowman, W.P.; George, S.L.; Pui, C.H. Clinical pharmacodynamics of high-dose methotrexate in acute lymphocytic leukemia. Identification of a relation between concentration and effect. N. Engl. J. Med. 1986, 314, 471–477. [Google Scholar] [CrossRef]
- Ferreri, A.J.; Reni, M.; Foppoli, M.; Martelli, M.; Pangalis, G.A.; Frezzato, M.; Cabras, M.G.; Fabbri, A.; Corazzelli, G.; Ilariucci, F.; et al. High-dose cytarabine plus high-dose methotrexate versus high-dose methotrexate alone in patients with primary CNS lymphoma: A randomised phase 2 trial. Lancet 2009, 374, 1512–1520. [Google Scholar] [CrossRef]
- Grem, J.L.; King, S.A.; Wittes, R.E.; Leyland-Jones, B. The role of methotrexate in osteosarcoma. J. Natl. Cancer Inst. 1988, 80, 626–655. [Google Scholar] [CrossRef]
- Kozminski, P.; Halik, P.K.; Chesori, R.; Gniazdowska, E. Overview of Dual-Acting Drug Methotrexate in Different Neurological Diseases, Autoimmune Pathologies and Cancers. Int. J. Mol. Sci. 2020, 21, 3483. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, P.T.; Zhang, Z.; McCourt, L.; Dwyer, M.; Benkovic, S.J.; Hammes, G.G. Interaction of dihydrofolate reductase with methotrexate: Ensemble and single-molecule kinetics. Proc. Natl. Acad. Sci. USA 2002, 99, 13481–13486. [Google Scholar] [CrossRef] [Green Version]
- Raimondi, M.V.; Randazzo, O.; La Franca, M.; Barone, G.; Vignoni, E.; Rossi, D.; Collina, S. DHFR Inhibitors: Reading the Past for Discovering Novel Anticancer Agents. Molecules 2019, 24, 1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pui, C.H.; Evans, W.E. Treatment of acute lymphoblastic leukemia. N. Engl. J. Med. 2006, 354, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Vader, J.P.; Burnand, B.; Froehlich, F.; Dubois, R.W.; Bochud, M.; Gonvers, J.J. The European Panel on Appropriateness of Gastrointestinal Endoscopy (EPAGE): Project and methods. Endoscopy 1999, 31, 572–578. [Google Scholar] [CrossRef]
- Strahlendorf, C.; Pole, J.D.; Barber, R.; Dix, D.; Kulkarni, K.; Martineau, E.; Randall, A.; Stammers, D.; Strother, D.; Truong, T.H.; et al. Enrolling children with acute lymphoblastic leukaemia on a clinical trial improves event-free survival: A population-based study. Br. J. Cancer 2018, 118, 744–749. [Google Scholar] [CrossRef] [Green Version]
- Widemann, B.C.; Adamson, P.C. Understanding and managing methotrexate nephrotoxicity. Oncologist 2006, 11, 694–703. [Google Scholar] [CrossRef] [Green Version]
- Howard, S.C.; McCormick, J.; Pui, C.H.; Buddington, R.K.; Harvey, R.D. Preventing and Managing Toxicities of High-Dose Methotrexate. Oncologist 2016, 21, 1471–1482. [Google Scholar] [CrossRef] [Green Version]
- Schornagel, J.H.; McVie, J.G. The clinical pharmacology of methotrexate. Cancer Treat. Rev. 1983, 10, 53–75. [Google Scholar] [CrossRef]
- Shen, D.D.; Azarnoff, D.L. Clinical Pharmacokinetics of Methotrexate. Clin. Pharmacokinet. 1978, 3, 1–13. [Google Scholar] [CrossRef]
- Alsdorf, W.H.; Karagiannis, P.; Langebrake, C.; Bokemeyer, C.; Frenzel, C. Standardized Supportive Care Documentation Improves Safety of High-Dose Methotrexate Treatment. Oncologist 2021, 26, e327–e332. [Google Scholar] [CrossRef]
- Taylor, Z.L.; Mizuno, T.; Punt, N.C.; Baskaran, B.; Navarro Sainz, A.; Shuman, W.; Felicelli, N.; Vinks, A.A.; Heldrup, J.; Ramsey, L.B. MTXPK.org: A Clinical Decision Support Tool Evaluating High-Dose Methotrexate Pharmacokinetics to Inform Post-Infusion Care and Use of Glucarpidase. Clin. Pharm. Ther. 2020, 108, 635–643. [Google Scholar] [CrossRef]
- Chanfreau-Coffinier, C.; Hull, L.E.; Lynch, J.A.; DuVall, S.L.; Damrauer, S.M.; Cunningham, F.E.; Voight, B.F.; Matheny, M.E.; Oslin, D.W.; Icardi, M.S.; et al. Projected Prevalence of Actionable Pharmacogenetic Variants and Level A Drugs Prescribed Among US Veterans Health Administration Pharmacy Users. JAMA Netw. Open 2019, 2, e195345. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.H.C.; Chan, M.C.Y.; Chung, C.C.Y.; Li, A.W.T.; Yip, C.Y.W.; Mak, C.C.Y.; Chau, J.F.T.; Lee, M.; Fung, J.L.F.; Tsang, M.H.Y.; et al. Actionable pharmacogenetic variants in Hong Kong Chinese exome sequencing data and projected prescription impact in the Hong Kong population. PLoS Genet. 2021, 17, e1009323. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegyi, M.; Arany, A.; Semsei, A.F.; Csordas, K.; Eipel, O.; Gezsi, A.; Kutszegi, N.; Csoka, M.; Muller, J.; Erdelyi, D.J.; et al. Pharmacogenetic analysis of high-dose methotrexate treatment in children with osteosarcoma. Oncotarget 2017, 8, 9388–9398. [Google Scholar] [CrossRef] [Green Version]
- Csordas, K.; Lautner-Csorba, O.; Semsei, A.F.; Harnos, A.; Hegyi, M.; Erdelyi, D.J.; Eipel, O.T.; Szalai, C.; Kovacs, G.T. Associations of novel genetic variations in the folate-related and ARID5B genes with the pharmacokinetics and toxicity of high-dose methotrexate in paediatric acute lymphoblastic leukaemia. Br. J. Haematol. 2014, 166, 410–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lui, G.; Treluyer, J.M.; Fresneau, B.; Piperno-Neumann, S.; Gaspar, N.; Corradini, N.; Gentet, J.C.; Marec Berard, P.; Laurence, V.; Schneider, P.; et al. A Pharmacokinetic and Pharmacogenetic Analysis of Osteosarcoma Patients Treated With High-Dose Methotrexate: Data From the OS2006/Sarcoma-09 Trial. J. Clin. Pharm. 2018, 58, 1541–1549. [Google Scholar] [CrossRef] [PubMed]
- Yanagimachi, M.; Goto, H.; Kaneko, T.; Naruto, T.; Sasaki, K.; Takeuchi, M.; Tanoshima, R.; Kato, H.; Yokosuka, T.; Kajiwara, R.; et al. Influence of pre-hydration and pharmacogenetics on plasma methotrexate concentration and renal dysfunction following high-dose methotrexate therapy. Int. J. Hematol. 2013, 98, 702–707. [Google Scholar] [CrossRef] [PubMed]
- El Mesallamy, H.O.; Rashed, W.M.; Hamdy, N.M.; Hamdy, N. High-dose methotrexate in Egyptian pediatric acute lymphoblastic leukemia: The impact of ABCG2 C421A genetic polymorphism on plasma levels, what is next? J. Cancer Res. Clin. Oncol. 2014, 140, 1359–1365. [Google Scholar] [CrossRef]
- Lopez-Lopez, E.; Martin-Guerrero, I.; Ballesteros, J.; Pinan, M.A.; Garcia-Miguel, P.; Navajas, A.; Garcia-Orad, A. Polymorphisms of the SLCO1B1 gene predict methotrexate-related toxicity in childhood acute lymphoblastic leukemia. Pediatric Blood Cancer 2011, 57, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Esmaili, M.A.; Kazemi, A.; Faranoush, M.; Mellstedt, H.; Zaker, F.; Safa, M.; Mehrvar, N.; Rezvany, M.R. Polymorphisms within methotrexate pathway genes: Relationship between plasma methotrexate levels, toxicity experienced and outcome in pediatric acute lymphoblastic leukemia. Iran. J. Basic Med. Sci. 2020, 23, 800–809. [Google Scholar] [CrossRef]
- Cheng, Y.; Chen, M.H.; Zhuang, Q.; Lin, B.J.; Chen, Y.Y.; Yang, L.; Liu, M.B.; Que, W.C.; Qiu, H.Q. Genetic factors involved in delayed methotrexate elimination in children with acute lymphoblastic leukemia. Pediatric Blood Cancer 2021, 68, e28858. [Google Scholar] [CrossRef] [PubMed]
- Den Hoed, M.A.; Lopez-Lopez, E.; te Winkel, M.L.; Tissing, W.; de Rooij, J.D.; Gutierrez-Camino, A.; Garcia-Orad, A.; den Boer, E.; Pieters, R.; Pluijm, S.M.; et al. Genetic and metabolic determinants of methotrexate-induced mucositis in pediatric acute lymphoblastic leukemia. Pharm. J. 2015, 15, 248–254. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Lopez, E.; Ballesteros, J.; Pinan, M.A.; Sanchez de Toledo, J.; Garcia de Andoin, N.; Garcia-Miguel, P.; Navajas, A.; Garcia-Orad, A. Polymorphisms in the methotrexate transport pathway: A new tool for MTX plasma level prediction in pediatric acute lymphoblastic leukemia. Pharm. Genom. 2013, 23, 53–61. [Google Scholar] [CrossRef]
- Rau, T.; Erney, B.; Gores, R.; Eschenhagen, T.; Beck, J.; Langer, T. High-dose methotrexate in pediatric acute lymphoblastic leukemia: Impact of ABCC2 polymorphisms on plasma concentrations. Clin. Pharm. Ther. 2006, 80, 468–476. [Google Scholar] [CrossRef]
- Liu, Y.; Yin, Y.; Sheng, Q.; Lu, X.; Wang, F.; Lin, Z.; Tian, H.; Xu, A.; Zhang, J. Association of ABCC2 -24C>T polymorphism with high-dose methotrexate plasma concentrations and toxicities in childhood acute lymphoblastic leukemia. PLoS ONE 2014, 9, e82681. [Google Scholar] [CrossRef] [PubMed]
- Razali, R.H.; Noorizhab, M.N.F.; Jamari, H.; James, R.J.; Teh, K.H.; Ibrahim, H.M.; Teh, L.K.; Salleh, M.Z. Association of ABCC2 with levels and toxicity of methotrexate in Malaysian Childhood Acute Lymphoblastic Leukemia (ALL). Pediatr. Hematol. Oncol. 2020, 37, 185–197. [Google Scholar] [CrossRef]
- Radtke, S.; Zolk, O.; Renner, B.; Paulides, M.; Zimmermann, M.; Moricke, A.; Stanulla, M.; Schrappe, M.; Langer, T. Germline genetic variations in methotrexate candidate genes are associated with pharmacokinetics, toxicity, and outcome in childhood acute lymphoblastic leukemia. Blood 2013, 121, 5145–5153. [Google Scholar] [CrossRef] [Green Version]
- Zgheib, N.K.; Akra-Ismail, M.; Aridi, C.; Mahfouz, R.; Abboud, M.R.; Solh, H.; Muwakkit, S.A. Genetic polymorphisms in candidate genes predict increased toxicity with methotrexate therapy in Lebanese children with acute lymphoblastic leukemia. Pharm. Genom. 2014, 24, 387–396. [Google Scholar] [CrossRef]
- Goricar, K.; Kovac, V.; Jazbec, J.; Zakotnik, B.; Lamovec, J.; Dolzan, V. Influence of the folate pathway and transporter polymorphisms on methotrexate treatment outcome in osteosarcoma. Pharm. Genom. 2014, 24, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Ansari, M.; Sauty, G.; Labuda, M.; Gagne, V.; Rousseau, J.; Moghrabi, A.; Laverdiere, C.; Sinnett, D.; Krajinovic, M. Polymorphism in multidrug resistance-associated protein gene 3 is associated with outcomes in childhood acute lymphoblastic leukemia. Pharm. J. 2012, 12, 386–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ansari, M.; Sauty, G.; Labuda, M.; Gagne, V.; Laverdiere, C.; Moghrabi, A.; Sinnett, D.; Krajinovic, M. Polymorphisms in multidrug resistance-associated protein gene 4 is associated with outcome in childhood acute lymphoblastic leukemia. Blood 2009, 114, 1383–1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.G.; Gao, C.; Zhang, R.D.; Zhao, X.X.; Cui, L.; Li, W.J.; Chen, Z.P.; Yue, Z.X.; Zhang, Y.Y.; Wu, M.Y.; et al. Polymorphisms in methotrexate transporters and their relationship to plasma methotrexate levels, toxicity of high-dose methotrexate, and outcome of pediatric acute lymphoblastic leukemia. Oncotarget 2017, 8, 37761–37772. [Google Scholar] [CrossRef] [PubMed]
- Imanishi, H.; Okamura, N.; Yagi, M.; Noro, Y.; Moriya, Y.; Nakamura, T.; Hayakawa, A.; Takeshima, Y.; Sakaeda, T.; Matsuo, M.; et al. Genetic polymorphisms associated with adverse events and elimination of methotrexate in childhood acute lymphoblastic leukemia and malignant lymphoma. J. Hum. Genet. 2007, 52, 166–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laverdiere, C.; Chiasson, S.; Costea, I.; Moghrabi, A.; Krajinovic, M. Polymorphism G80A in the reduced folate carrier gene and its relationship to methotrexate plasma levels and outcome of childhood acute lymphoblastic leukemia. Blood 2002, 100, 3832–3834. [Google Scholar] [CrossRef]
- Gregers, J.; Christensen, I.J.; Dalhoff, K.; Lausen, B.; Schroeder, H.; Rosthoej, S.; Carlsen, N.; Schmiegelow, K.; Peterson, C. The association of reduced folate carrier 80G>A polymorphism to outcome in childhood acute lymphoblastic leukemia interacts with chromosome 21 copy number. Blood 2010, 115, 4671–4677. [Google Scholar] [CrossRef] [Green Version]
- Cwiklinska, M.; Czogala, M.; Kwiecinska, K.; Madetko-Talowska, A.; Szafarz, M.; Pawinska, K.; Wieczorek, A.; Klekawka, T.; Rej, M.; Stepien, K.; et al. Polymorphisms of SLC19A1 80 G>A, MTHFR 677 C>T, and Tandem TS Repeats Influence Pharmacokinetics, Acute Liver Toxicity, and Vomiting in Children With Acute Lymphoblastic Leukemia Treated With High Doses of Methotrexate. Front. Pediatr. 2020, 8, 307. [Google Scholar] [CrossRef]
- Kotur, N.; Lazic, J.; Ristivojevic, B.; Stankovic, B.; Gasic, V.; Dokmanovic, L.; Krstovski, N.; Milosevic, G.; Janic, D.; Zukic, B.; et al. Pharmacogenomic Markers of Methotrexate Response in the Consolidation Phase of Pediatric Acute Lymphoblastic Leukemia Treatment. Genes 2020, 11, 468. [Google Scholar] [CrossRef]
- Wang, S.M.; Sun, L.L.; Zeng, W.X.; Wu, W.S.; Zhang, G.L. Effects of a microRNA binding site polymorphism in SLC19A1 on methotrexate concentrations in Chinese children with acute lymphoblastic leukemia. Med. Oncol. 2014, 31, 62. [Google Scholar] [CrossRef] [PubMed]
- Park, J.A.; Shin, H.Y. Influence of genetic polymorphisms in the folate pathway on toxicity after high-dose methotrexate treatment in pediatric osteosarcoma. Blood Res. 2016, 51, 50–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishi, S.; Cheng, C.; French, D.; Pei, D.; Das, S.; Cook, E.H.; Hijiya, N.; Rizzari, C.; Rosner, G.L.; Frudakis, T.; et al. Ancestry and pharmacogenetics of antileukemic drug toxicity. Blood 2007, 109, 4151–4157. [Google Scholar] [CrossRef]
- Shimasaki, N.; Mori, T.; Samejima, H.; Sato, R.; Shimada, H.; Yahagi, N.; Torii, C.; Yoshihara, H.; Tanigawara, Y.; Takahashi, T.; et al. Effects of methylenetetrahydrofolate reductase and reduced folate carrier 1 polymorphisms on high-dose methotrexate-induced toxicities in children with acute lymphoblastic leukemia or lymphoma. J. Pediatr. Hematol. Oncol. 2006, 28, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.M.; Zeng, W.X.; Wu, W.S.; Sun, L.L.; Yan, D. Association between a microRNA binding site polymorphism in SLCO1A2 and the risk of delayed methotrexate elimination in Chinese children with acute lymphoblastic leukemia. Leuk. Res. 2018, 65, 61–66. [Google Scholar] [CrossRef]
- Trevino, L.R.; Shimasaki, N.; Yang, W.; Panetta, J.C.; Cheng, C.; Pei, D.; Chan, D.; Sparreboom, A.; Giacomini, K.M.; Pui, C.H.; et al. Germline genetic variation in an organic anion transporter polypeptide associated with methotrexate pharmacokinetics and clinical effects. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 5972–5978. [Google Scholar] [CrossRef] [Green Version]
- Ramsey, L.B.; Panetta, J.C.; Smith, C.; Yang, W.; Fan, Y.; Winick, N.J.; Martin, P.L.; Cheng, C.; Devidas, M.; Pui, C.H.; et al. Genome-wide study of methotrexate clearance replicates SLCO1B1. Blood 2013, 121, 898–904. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, L.B.; Bruun, G.H.; Yang, W.; Trevino, L.R.; Vattathil, S.; Scheet, P.; Cheng, C.; Rosner, G.L.; Giacomini, K.M.; Fan, Y.; et al. Rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res. 2012, 22, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Fukushima, H.; Fukushima, T.; Sakai, A.; Suzuki, R.; Nakajima-Yamaguchi, R.; Kobayashi, C.; Iwabuchi, A.; Saito, M.; Yoshimi, A.; Nakao, T.; et al. Polymorphisms of MTHFR Associated with Higher Relapse/Death Ratio and Delayed Weekly MTX Administration in Pediatric Lymphoid Malignancies. Leuk. Res. Treat. 2013, 2013, 238528. [Google Scholar] [CrossRef]
- Li, J.; Wang, X.R.; Zhai, X.W.; Wang, H.S.; Qian, X.W.; Miao, H.; Zhu, X.H. Association of SLCO1B1 gene polymorphisms with toxicity response of high dose methotrexate chemotherapy in childhood acute lymphoblastic leukemia. Int. J. Clin. Exp. Med. 2015, 8, 6109–6113. [Google Scholar] [PubMed]
- Schulte, R.R.; Choi, L.; Utreja, N.; Van Driest, S.L.; Stein, C.M.; Ho, R.H. Effect of SLCO1B1 Polymorphisms on High-Dose Methotrexate Clearance in Children and Young Adults With Leukemia and Lymphoblastic Lymphoma. Clin. Transl. Sci. 2021, 14, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.N.; He, X.L.; Wang, C.; Wang, Y.; Chen, Y.J.; Li, J.X.; Niu, C.H.; Gao, P. Impact of SLCO1B1 521T > C variant on leucovorin rescue and risk of relapse in childhood acute lymphoblastic leukemia treated with high-dose methotrexate. Pediatric Blood Cancer 2014, 61, 2203–2207. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Faganel Kotnik, B.; Grabnar, I.; Bohanec Grabar, P.; Dolzan, V.; Jazbec, J. Association of genetic polymorphism in the folate metabolic pathway with methotrexate pharmacokinetics and toxicity in childhood acute lymphoblastic leukaemia and malignant lymphoma. Eur. J. Clin. Pharm. 2011, 67, 993–1006. [Google Scholar] [CrossRef]
- Race, J.E.; Grassl, S.M.; Williams, W.J.; Holtzman, E.J. Molecular cloning and characterization of two novel human renal organic anion transporters (hOAT1 and hOAT3). Biochem. Biophys. Res. Commun. 1999, 255, 508–514. [Google Scholar] [CrossRef]
- Liu, Z.H.; Jia, Y.M.; Wang, C.Y.; Meng, Q.; Huo, X.K.; Sun, H.J.; Sun, P.Y.; Yang, X.B.; Ma, X.D.; Peng, J.Y.; et al. Organic anion transporters 1 (OAT1) and OAT3 meditated the protective effect of rhein on methotrexate-induced nephrotoxicity. RSC Adv. 2017, 7, 25461–25468. [Google Scholar] [CrossRef] [Green Version]
- Chioukh, R.; Noel-Hudson, M.S.; Ribes, S.; Fournier, N.; Becquemont, L.; Verstuyft, C. Proton pump inhibitors inhibit methotrexate transport by renal basolateral organic anion transporter hOAT3. Drug Metab. Dispos. Biol. Fate Chem. 2014, 42, 2041–2048. [Google Scholar] [CrossRef] [Green Version]
- Cha, S.H.; Sekine, T.; Fukushima, J.I.; Kanai, Y.; Kobayashi, Y.; Goya, T.; Endou, H. Identification and characterization of human organic anion transporter 3 expressing predominantly in the kidney. Mol. Pharmacol. 2001, 59, 1277–1286. [Google Scholar] [CrossRef] [PubMed]
- Erdman, A.R.; Mangravite, L.M.; Urban, T.J.; Lagpacan, L.L.; Castro, R.A.; de la Cruz, M.; Chan, W.; Huang, C.C.; Johns, S.J.; Kawamoto, M.; et al. The human organic anion transporter 3 (OAT3; SLC22A8): Genetic variation and functional genomics. Am. J. Physiol. Ren. Physiol. 2006, 290, F905–F912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VanWert, A.L.; Sweet, D.H. Impaired clearance of methotrexate in organic anion transporter 3 (Slc22a8) knockout mice: A gender specific impact of reduced folates. Pharm. Res. 2008, 25, 453–462. [Google Scholar] [CrossRef] [Green Version]
- Tirona, R.G.; Leake, B.F.; Merino, G.; Kim, R.B. Polymorphisms in OATP-C: Identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J. Biol. Chem. 2001, 276, 35669–35675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nies, A.T.; Niemi, M.; Burk, O.; Winter, S.; Zanger, U.M.; Stieger, B.; Schwab, M.; Schaeffeler, E. Genetics is a major determinant of expression of the human hepatic uptake transporter OATP1B1, but not of OATP1B3 and OATP2B1. Genome Med. 2013, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Chae, Y.J.; Lee, K.R.; Noh, C.K.; Chong, S.; Kim, D.D.; Shim, C.K.; Chung, S.J. Functional consequences of genetic variations in the human organic anion transporting polypeptide 1B3 (OATP1B3) in the Korean population. J. Pharm. Sci. 2012, 101, 1302–1313. [Google Scholar] [CrossRef]
- Wang, S.M.; Sun, L.L.; Zeng, W.X.; Wu, W.S.; Zhang, G.L. Influence of genetic polymorphisms of FPGS, GGH, and MTHFR on serum methotrexate levels in Chinese children with acute lymphoblastic leukemia. Cancer Chemother. Pharm. 2014, 74, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, S.M.; Wu, W.S.; Yan, D.; Zhang, L.P.; Zheng, H.Y. Frequency distribution of five SNPs in human GGH gene and their effects on clinical outcomes of Chinese pediatric patients with acute lymphoblastic leukemia. Die Pharm. 2020, 75, 142–146. [Google Scholar] [CrossRef]
- Huang, Z.; Tong, H.F.; Qian, J.C.; Wang, J.X.; Li, Y.; Chen, M.; Luan, Z. Association of folypolyglutamate synthetase (FPGS) gene polymorphism with blood drug concentration as well as adverse reactions of methotrexate in children with acute leukaemia. Biomed. Res. India 2017, 28, 478–483. [Google Scholar]
- Huang, Z.; Tong, H.F.; Li, Y.; Qian, J.C.; Wang, J.X.; Wang, Z.; Ruan, J.C. Effect of the Polymorphism of Folylpolyglutamate Synthetase on Treatment of High-Dose Methotrexate in Pediatric Patients with Acute Lymphocytic Leukemia. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2016, 22, 4967–4973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erculj, N.; Kotnik, B.F.; Debeljak, M.; Jazbec, J.; Dolzan, V. Influence of folate pathway polymorphisms on high-dose methotrexate-related toxicity and survival in childhood acute lymphoblastic leukemia. Leuk. Lymphoma 2012, 53, 1096–1104. [Google Scholar] [CrossRef]
- Erculj, N.; Kotnik, B.F.; Debeljak, M.; Jazbec, J.; Dolzan, V. The influence of folate pathway polymorphisms on high-dose methotrexate-related toxicity and survival in children with non-Hodgkin malignant lymphoma. Radiol. Oncol. 2014, 48, 289–292. [Google Scholar] [CrossRef] [Green Version]
- Mahmoud, L.B.; Mdhaffar, M.; Frikha, R.; Ghozzi, H.; Hakim, A.; Sahnoun, Z.; Elloumi, M.; Zeghal, K. Use of MTHFR C677T polymorphism and plasma pharmacokinetics to predict methotrexate toxicity in patients with acute lymphoblastic leukemia. Adv. Clin. Exp. Med. Off. Organ Wroc. Med Univ. 2018, 27, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Park, H.; Lee, J.S.; Lee, J.Y.; Kim, S.; Kim, T.W.; Park, J.S.; Kim, J.E.; Yoon, D.H.; Suh, C. Methotrexate elimination and toxicity: MTHFR 677C>T polymorphism in patients with primary CNS lymphoma treated with high-dose methotrexate. Hematol. Oncol. 2017, 35, 504–509. [Google Scholar] [CrossRef]
- Yousef, A.M.; Farhad, R.; Alshamaseen, D.; Alsheikh, A.; Zawiah, M.; Kadi, T. Folate pathway genetic polymorphisms modulate methotrexate-induced toxicity in childhood acute lymphoblastic leukemia. Cancer Chemother. Pharm. 2019, 83, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Kaluzna, E.; Strauss, E.; Zajac-Spychala, O.; Gowin, E.; Swiatek-Koscielna, B.; Nowak, J.; Fichna, M.; Mankowski, P.; Januszkiewicz-Lewandowska, D. Functional variants of gene encoding folate metabolizing enzyme and methotrexate-related toxicity in children with acute lymphoblastic leukemia. Eur. J. Pharm. 2015, 769, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, L.; Sleurs, C.; Labarque, V.; Dhooge, C.; Laenen, A.; Sinnaeve, F.; Renard, M.; Uyttebroeck, A. The role of the MTHFR C677T polymorphism in methotrexate-induced toxicity in pediatric osteosarcoma patients. Pharmacogenomics 2017, 18, 787–795. [Google Scholar] [CrossRef]
- El-Khodary, N.M.; El-Haggar, S.M.; Eid, M.A.; Ebeid, E.N. Study of the pharmacokinetic and pharmacogenetic contribution to the toxicity of high-dose methotrexate in children with acute lymphoblastic leukemia. Med. Oncol. 2012, 29, 2053–2062. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.G.; Li, Z.G.; Cui, L.; Gao, C.; Li, W.J.; Zhao, X.X. Effects of methylenetetrahydrofolate reductase gene polymorphisms on toxicities during consolidation therapy in pediatric acute lymphoblastic leukemia in a Chinese population. Leuk. Lymphoma 2011, 52, 1030–1040. [Google Scholar] [CrossRef]
- De Deus, D.M.; de Lima, E.L.; Seabra Silva, R.M.; Leite, E.P.; Cartaxo Muniz, M.T. Influence of Methylenetetrahydrofolate Reductase C677T, A1298C, and G80A Polymorphisms on the Survival of Pediatric Patients with Acute Lymphoblastic Leukemia. Leuk. Res. Treat. 2012, 2012, 292043. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.M.; Zeng, W.X.; Wu, W.S.; Sun, L.L.; Yan, D. Genotype and allele frequencies of TYMS rs2790 A > G polymorphism in a Chinese paediatric population with acute lymphoblastic leukaemia. J. Clin. Pharm. Ther. 2018, 43, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Deng, Y.; Yan, X.; Zhu, J.; Yin, Y.; Shu, Y.; Bai, D.; Zhang, S.; Xu, H.; Lu, X. The Role of ARID5B in Acute Lymphoblastic Leukemia and Beyond. Front. Genet. 2020, 11, 598. [Google Scholar] [CrossRef]
- Wang, S.M.; Kong, X.Y.; Li, M.; Sun, L.L.; Yan, D. Association of GGH Promoter Methylation Levels with Methotrexate Concentrations in Chinese Children with Acute Lymphoblastic Leukemia. Pharmacotherapy 2020, 40, 614–622. [Google Scholar] [CrossRef] [PubMed]
- Krajinovic, M. MTHFD1 gene: Role in disease susceptibility and pharmacogenetics. Pharmacogenomics 2008, 9, 829–832. [Google Scholar] [CrossRef] [PubMed]
- Windsor, R.E.; Strauss, S.J.; Kallis, C.; Wood, N.E.; Whelan, J.S. Germline genetic polymorphisms may influence chemotherapy response and disease outcome in osteosarcoma: A pilot study. Cancer 2012, 118, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Lopez, E.; Martin-Guerrero, I.; Ballesteros, J.; Garcia-Orad, A. A systematic review and meta-analysis of MTHFR polymorphisms in methotrexate toxicity prediction in pediatric acute lymphoblastic leukemia. Pharm. J. 2013, 13, 498–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horie, N.; Aiba, H.; Oguro, K.; Hojo, H.; Takeishi, K. Functional analysis and DNA polymorphism of the tandemly repeated sequences in the 5′-terminal regulatory region of the human gene for thymidylate synthase. Cell Struct. Funct. 1995, 20, 191–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorich, M.J.; Pottier, N.; Pei, D.; Yang, W.; Kager, L.; Stocco, G.; Cheng, C.; Panetta, J.C.; Pui, C.H.; Relling, M.V.; et al. In vivo response to methotrexate forecasts outcome of acute lymphoblastic leukemia and has a distinct gene expression profile. PLoS Med. 2008, 5, e83. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.J.; Cheng, C.; Yang, W.; Pei, D.; Cao, X.; Fan, Y.; Pounds, S.B.; Neale, G.; Trevino, L.R.; French, D.; et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA 2009, 301, 393–403. [Google Scholar] [CrossRef] [Green Version]
- Hurkmans, E.G.E.; Klumpers, M.J.; Vermeulen, S.H.; Hagleitner, M.M.; Flucke, U.; Schreuder, H.W.B.; Gelderblom, H.; Bras, J.; Guchelaar, H.J.; Coenen, M.J.H.; et al. Analysis of Drug Metabolizing Gene Panel in Osteosarcoma Patients Identifies Association Between Variants in SULT1E1, CYP2B6 and CYP4F8 and Methotrexate Levels and Toxicities. Front. Pharm. 2020, 11, 1241. [Google Scholar] [CrossRef]
- Van de Steeg, E.; van der Kruijssen, C.M.M.; Wagenaar, E.; Burggraaff, J.E.C.; Mesman, E.; Kenworthy, K.E.; Schinkel, A.H. Methotrexate Pharmacokinetics in Transgenic Mice with Liver-Specific Expression of Human Organic Anion-Transporting Polypeptide 1B1 (SLCO1B1). Drug Metab. Dispos. 2009, 37, 277–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlaming, M.L.H.; Pala, Z.; van Esch, A.; Wagenaar, E.; van Tellingen, O.; de Waart, D.R.; Oude Elferink, R.P.J.; van de Wetering, K.; Schinkel, A.H. Impact of Abcc2 (Mrp2) and Abcc3 (Mrp3) on the In vivo Elimination of Methotrexate and its Main Toxic Metabolite 7-hydroxymethotrexate. Clin. Cancer Res. 2008, 14, 8152–8160. [Google Scholar] [CrossRef] [Green Version]
- Muller, J.; Kralovanszky, J.; Adleff, V.; Pap, E.; Nemeth, K.; Komlosi, V.; Kovacs, G. Toxic encephalopathy and delayed MTX clearance after high-dose methotrexate therapy in a child homozygous for the MTHFR C677T polymorphism. Anticancer Res. 2008, 28, 3051–3054. [Google Scholar] [PubMed]
- Hulot, J.-S.; Villard, E.; Maguy, A.; Morel, V.; Mir, L.; Tostivint, I.; William-Faltaos, D.; Fernandez, C.; Hatem, S.; Deray, G.; et al. A mutation in the drug transporter gene ABCC2 associated with impaired methotrexate elimination. Pharm. Genom. 2005, 15, 277–285. [Google Scholar] [CrossRef]
- Gaedigk, A.; Sangkuhl, K.; Whirl-Carrillo, M.; Twist, G.P.; Klein, T.E.; Miller, N.A. The Evolution of PharmVar. Clin. Pharm. Ther. 2019, 105, 29–32. [Google Scholar] [CrossRef] [Green Version]
- Taparia, S.; Gelineau-van Waes, J.; Rosenquist, T.H.; Finnell, R.H. Importance of folate-homocysteine homeostasis during early embryonic development. Clin. Chem. Lab. Med. 2007, 45, 1717–1727. [Google Scholar] [CrossRef]
- Ioannidis, J.P.; Trikalinos, T.A.; Khoury, M.J. Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. Am. J. Epidemiol. 2006, 164, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Cho, J.; Zhao, H. Practical issues in building risk-predicting models for complex diseases. J. Biopharm. Stat. 2010, 20, 415–440. [Google Scholar] [CrossRef]
- Nicholls, H.L.; John, C.R.; Watson, D.S.; Munroe, P.B.; Barnes, M.R.; Cabrera, C.P. Reaching the End-Game for GWAS: Machine Learning Approaches for the Prioritization of Complex Disease Loci. Front. Genet. 2020, 11, 350. [Google Scholar] [CrossRef]
- Pérez-Enciso, M.; Zingaretti, L.M. A Guide for Using Deep Learning for Complex Trait Genomic Prediction. Genes 2019, 10, 553. [Google Scholar] [CrossRef] [Green Version]



| Gene Name | Variants | Effect of Variant on Methotrexate Exposure | References |
|---|---|---|---|
| ABCB1 | rs9282564 | ↑ | [20,21] |
| ABCG2 | rs12505410 | ↓ | [22] |
| ABCG2 | rs13120400 | ↑ | [22] |
| ABCG2 | rs13137622 | ↓ | [22] |
| ABCG2 | rs2231142 | ↑ | [20,23,24,25,26,27] |
| ABCC2 | rs3740065 | ↑ *|↓ | [22,28,29*,30*,31*] |
| ABCC2 | rs3740066 | ↑ *|↓ | [20,27*,29*] |
| ABCC2 | rs717620 | ↑ *|↓ | [20,22,23,27,28,29,30*,31*,32,33,34,35,36] |
| ABCC3 | rs4793665 | ↓ | [20,36] |
| ABCC3 | rs9895420 | ↓ | [36] |
| ABCC4 | rs10219913 | ↑ | [28,29] |
| ABCC4 | rs7317112 | ↑ | [28,29] |
| ABCC4 | rs868853 | ↓ | [29,31,37] |
| ABCC4 | rs9516519 | ↑ *|↓ | [28*,29,31*] |
| SLC19A1 | rs1051266 | ↑ *|↓ | [20,21,22,26*,27,28,29*,33,34,35,38,39,40*,41*,42*,43*] |
| SLC19A1 | rs1051296 | ↑ | [44] |
| SLC19A1 | rs61510559 | ↓ | [23,25,45,46,47] |
| SLCO1A2 | rs4149009 | ↓ | [27,29,48] |
| SLCO1B1 | rs10841753 | ↑ *|↓ | [38,49*,50] |
| SLCO1B1 | rs11045787 | ↑ | [49] |
| SLCO1B1 | rs11045818 | ↑ *|↓ | [21,49*,50*] |
| SLCO1B1 | rs11045813 | ↑ | [50] |
| SLCO1B1 | rs11045819 | ↓ | [21,51] |
| SLCO1B1 | rs11045821 | ↓ | [50] |
| SLCO1B1 | rs11045825 | ↓ | [50] |
| SLCO1B1 | rs11045870 | ↓ | [50] |
| SLCO1B1 | rs11045872 | ↓ | [22,49,50] |
| SLCO1B1 | rs11045879 | ↑ *|↓ | [22,25*,29,33,35*,49,50*,52] |
| SLCO1B1 | rs11045892 | ↓ | [50] |
| SLCO1B1 | rs11045897 | ↑ | [38,53] |
| SLCO1B1 | rs16923647 | ↓ | [50] |
| SLCO1B1 | rs17328763 | ↑ | [49] |
| SLCO1B1 | rs2169969 | ↓ | [50] |
| SLCO1B1 | rs2306283 | ↑ *|↓ | [27*,33,38,54] |
| SLCO1B1 | rs2900476 | ↓ | [22,49] |
| SLCO1B1 | rs34671512 | ↓ | [51] |
| SLCO1B1 | rs4149056 | ↑ | [21,22,27,28,32,33,34,35,38,43,49,50,51,52,54,55] |
| SLCO1B1 | rs4149076 | ↓ | [49] |
| SLCO1B1 | rs4149081 | ↑ *|↓ | [22*,49,50*,25*,53*] |
| SLCO1B1 | rs59502379 | ↑ | [51] |
| Gene Name | Variants | Effect of Variant on Methotrexate Exposure | References |
|---|---|---|---|
| ARID5B | rs4948496 | ↑ | [21,32] |
| GGH | rs3758149 | ↑ | [20,67,68] |
| FPGS | rs1544105 | ↓ | [69,70,67,21] |
| MTHFD1 | rs2236225 | ↓ | [21,35,71] |
| MTHFR | rs1801133 | ↑|↓ * | [23,25,26,27,28,32,33,34,35,39,42*,43,45,46,47,52,57,67,71*,72,73,74,75,76,77,78,79,80] |
| TYMS | rs2790 | ↓ | [81] |
| TYMS | rs34743033 | ↓ | [25,33,34,42,43,46,57,71] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taylor, Z.L.; Vang, J.; Lopez-Lopez, E.; Oosterom, N.; Mikkelsen, T.; Ramsey, L.B. Systematic Review of Pharmacogenetic Factors That Influence High-Dose Methotrexate Pharmacokinetics in Pediatric Malignancies. Cancers 2021, 13, 2837. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13112837
Taylor ZL, Vang J, Lopez-Lopez E, Oosterom N, Mikkelsen T, Ramsey LB. Systematic Review of Pharmacogenetic Factors That Influence High-Dose Methotrexate Pharmacokinetics in Pediatric Malignancies. Cancers. 2021; 13(11):2837. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13112837
Chicago/Turabian StyleTaylor, Zachary L., Jesper Vang, Elixabet Lopez-Lopez, Natanja Oosterom, Torben Mikkelsen, and Laura B. Ramsey. 2021. "Systematic Review of Pharmacogenetic Factors That Influence High-Dose Methotrexate Pharmacokinetics in Pediatric Malignancies" Cancers 13, no. 11: 2837. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13112837