Blockade LAT1 Mediates Methionine Metabolism to Overcome Oxaliplatin Resistance under Hypoxia in Renal Cell Carcinoma
Abstract
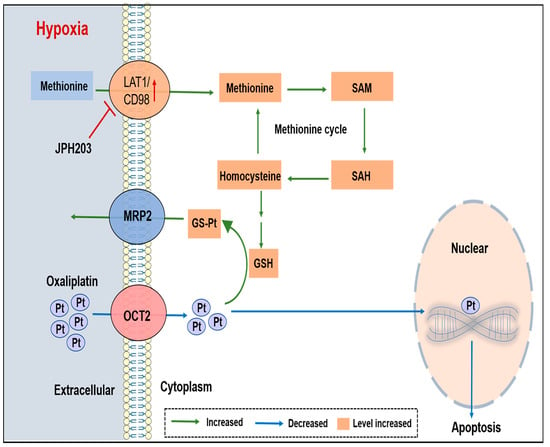
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Drug Treatment
2.2. Clinical Samples Collection
2.3. Real-Time Quantitative PCR and Western Blotting
2.4. Metabolomics Analysis
2.5. Construction of Knockdown and Overexpression RCC Cells
2.6. Reduced Glutathione Assays
2.7. Platinum Quantification
2.8. Cell Viability Assay
2.9. Animal Studies
2.10. Statistical Analyses
3. Results
3.1. Methionine Metabolism Is Induced in RCC Cells under Hypoxic Condition
3.2. Upregulated LAT1 Mediates Methionine Accumulation in RCC Cells under Hypoxia
3.3. LAT1 Could Be an Indicator for Grades and Overall Survival of RCC
3.4. LAT1 Mediates Oxaliplatin Accumulation in RCC Cells by Inhibiting GSH Production
3.5. LAT1 Inhibitor Combination with Oxaliplatin Potentiates Antitumor Efficiency In Vivo
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ljungberg, B.; Bensalah, K.; Canfield, S.; Dabestani, S.; Hofmann, F.; Hora, M.; Kuczyk, M.A.; Lam, T.; Marconi, L.; Merseburger, A.S.; et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur. Urol. 2015, 67, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Choueiri, T.K.; Motzer, R.J. Systemic therapy for metastatic renal cell carcinoma. N. Engl. J. Med. 2017, 376, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Barata, P.C.; Rini, B.I. Treatment of renal cell carcinoma: Current status and future directions. CA Cancer J. Clin. 2017, 67, 507–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nerich, V.; Hugues, M.; Paillard, M.J.; Borowski, L.; Nai, T.; Stein, U.; Nguyen Tan Hon, T.; Montcuquet, P.; Maurina, T.; Mouillet, G.; et al. Clinical impact of targeted therapies in patients with metastatic clear-cell renal cell carcinoma. OncoTargets Ther. 2014, 7, 365–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escudier, B. Combination therapy as first-line treatment in metastatic renal-cell carcinoma. N. Engl. J. Med. 2019, 380, 1176–1178. [Google Scholar] [CrossRef]
- Heidegger, I.; Pircher, A.; Pichler, R. Targeting the tumor microenvironment in renal cell cancer biology and therapy. Front. Radiat. Oncol. 2019, 9, 490. [Google Scholar] [CrossRef] [Green Version]
- Simonaggio, A.; Epaillard, N.; Pobel, C.; Moreira, M.; Oudard, S.; Vano, Y.A. Tumor microenvironment features as predictive biomarkers of response to immune checkpoint inhibitors (ICI) in metastatic clear cell renal cell carcinoma (mccRCC). Cancers 2021, 13, 231. [Google Scholar] [CrossRef]
- Vaupel, P.; Mayer, A. Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 2007, 26, 225–239. [Google Scholar] [CrossRef]
- Van den Borne, S.W.M.; Diez, J.; Blankesteijn, W.M.; Verjans, J.; Hofstra, L.; Narula, J. Myocardial remodeling after infarction: The role of myofibroblasts. Nat. Rev. Cardiol. 2010, 7, 30–37. [Google Scholar] [CrossRef]
- Ramachandran, S.; Ient, J.; Göttgens, E.L.; Krieg, A.J.; Hammond, E.M. Epigenetic therapy for solid tumors: Highlighting the impact of tumor hypoxia. Genes 2015, 6, 935–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wigerup, C.; Påhlman, S.; Bexell, D. Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer. Pharmacol. Ther. 2016, 164, 152–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warfel, N.A.; El Deiry, W.S. HIF-1 signaling in drug resistance to chemotherapy. Curr. Med. Chem. 2014, 21, 3021–3028. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Meierhofer, D. Glutathione metabolism in renal cell carcinoma progression and implications for therapies. Int. J. Mol. Sci. 2019, 20, 3672. [Google Scholar] [CrossRef] [Green Version]
- Wettersten, H.I.; Hakimi, A.A.; Morin, D.; Bianchi, C.; Johnstone, M.E.; Donohoe, D.R.; Trott, J.F.; Aboud, O.A.; Stirdivant, S.; Neri, B.; et al. Grade-dependent metabolic reprogramming in kidney cancer revealed by combined proteomics and metabolomics analysis. Cancer Res. 2015, 75, 2541–2552. [Google Scholar] [CrossRef] [Green Version]
- Wettersten, H.I.; Aboud, O.A.; Lara, P.N.; Weiss, R.H. Metabolic reprogramming in clear cell renal cell carcinoma. Nat. Rev. Nephrol. 2017, 13, 410–419. [Google Scholar] [CrossRef]
- Lauinger, L.; Kaiser, P. Sensing and signaling of methionine metabolism. Metabolites 2021, 11, 83. [Google Scholar] [CrossRef]
- Wang, X.; Guo, X.; Yu, W.; Li, C.; Gui, Y.; Cai, Z. Expression of methionine adenosyltransferase 2A in renal cell carcinomas and potential mechanism for kidney carcinogenesis. BMC Cancer 2014, 14, 196. [Google Scholar] [CrossRef] [Green Version]
- Dai, Z.; Mentch, S.J.; Gao, X.; Nichenametla, S.N.; Locasale, J.W. Methionine metabolism influences genomic architecture and gene expression through H3K4me3 peak width. Nat. Commun. 2018, 9, 1955. [Google Scholar] [CrossRef]
- Gu, X.; Orozco, J.M.; Saxton, R.A.; Condon, K.J.; Liu, G.Y.; Krawczyk, P.A.; Scaria, S.M.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 2017, 358, 813–818. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Bauer, C.; Newman, A.C.; Uribe, A.H.; Athineos, D.; Blyth, K.; Maddocks, O.D.K. Polyamine pathway activity promotes cysteine essentiality in cancer cells. Nat. Metab. 2020, 2, 1062–1076. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yip, L.Y.; Lee, J.H.J.; Wu, Z.; Chew, H.Y.; Chong, P.K.W.; Teo, C.C.; Ang, H.Y.; Peh, K.L.E.; Yuan, J.; et al. Methionine is a metabolic dependency of tumor-initiating cells. Nat. Med. 2019, 25, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.G.; Chen, J.; Mamane, V.; Ma, E.H.; Muhire, B.M.; Sheldon, R.D.; Shorstova, T.; Koning, R.; Johnson, R.M.; Esaulova, E.; et al. Methionine metabolism shapes T helper cell responses through regulation of epigenetic reprogramming. Cell Metab. 2020, 31, 250–266. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Sanderson, S.M.; Dai, Z.; Reid, M.A.; Cooper, D.E.; Lu, M.; Richie, J.P.; Ciccarella, A.; Calcagnotto, A.; Mikhael, P.G.; et al. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 2019, 572, 397–401. [Google Scholar] [CrossRef]
- Wanders, D.; Hobson, K.; Ji, X. Methionine restriction and cancer biology. Nutrients 2020, 12, 684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, M.H.; Lee, J.S.; Ma, C.; Diggs, L.P.; Heinrich, S.; Chang, C.W.; Ma, L.; Forgues, M.; Budhu, A.; Chaisaingmongkol, J.; et al. Tumor methionine metabolism drives T-cell exhaustion in hepatocellular carcinoma. Nat. Commun. 2021, 12, 1455. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Z.; Xu, Q.; Liu, Y.; Chen, L.; Guo, S.; Wang, H.; Zeng, K.; Liu, J.; Zeng, S.; et al. The failure of DAC to induce OCT2 expression and its remission by hemoglobin-based nanocarriers under hypoxia in renal cell carcinoma. Theranostics 2020, 10, 3562–3578. [Google Scholar] [CrossRef]
- Yan, R.; Zhao, X.; Lei, J.; Zhou, Q. Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex. Nature 2019, 568, 127–130. [Google Scholar] [CrossRef]
- Schödel, J.; Grampp, S.; Maher, E.R.; Moch, H.; Ratcliffe, P.J.; Russo, P.; Mole, D.R. Hypoxia, hypoxia-inducible transcription factors, and renal cancer. Eur. Urol. 2016, 69, 646–657. [Google Scholar] [CrossRef] [Green Version]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Lévi, F.; Metzger, G.; Massari, C.; Milano, G. Oxaliplatin: Pharmacokinetics and chronopharmacological aspects. Clin. Pharmacokinet. 2000, 38, 1–21. [Google Scholar] [PubMed]
- Jong, N.N.; Nakanishi, T.; Liu, J.J.; Tamai, I.; McKeage, M.J. Oxaliplatin transport mediated by organic cation/carnitine transporters OCTN1 and OCTN2 in overexpressing human embryonic kidney 293 cells and rat dorsal root ganglion neurons. J. Pharmacol. Exp. Ther. 2011, 338, 537–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrissey, K.M.; Wen, C.C.; Johns, S.J.; Zhang, L.; Huang, S.M.; Giacomini, K.M. The UCSF-FDA transportal: A public drug transporter database. Int. J. Clin. Pharmacol. Ther. 2012, 92, 545–546. [Google Scholar] [CrossRef] [Green Version]
- Martínez, Y.; Li, X.; Liu, G.; Bin, P.; Yan, W.; Más, D.; Valdivié, M.; Hu, C.A.A.; Ren, W.; Yin, Y. The role of methionine on metabolism, oxidative stress, and diseases. Amino Acids 2017, 49, 2091–2098. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, T.; Wang, J.; Li, T.W.H.; Fan, W.; Peng, H.; Krishnan, A.; Gores, G.J.; Mato, J.M.; Lu, S.C. Deregulated methionine adenosyltransferase α1, c-Myc, and Maf proteins together promote cholangiocarcinoma growth in mice and humans(‡). Hepatology 2016, 64, 439–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, H.; Tan, Y.; Kubota, T.; Moossa, A.R.; Hoffman, R.M. Methionine depletion modulates the antitumor and antimetastatic efficacy of ethionine. Anticancer Res. 1996, 16, 2719–2723. [Google Scholar]
- Sugisawa, N.; Hamada, K.; Han, Q.; Yamamoto, J.; Sun, Y.U.; Nishino, H.; Kawaguchi, K.; Bouvet, M.; Unno, M.; Hoffman, R.M. Adjuvant oral recombinant methioninase inhibits lung metastasis in a surgical breast cancer orthotopic syngeneic model. Anticancer Res. 2020, 40, 4869–4874. [Google Scholar] [CrossRef]
- Miousse, I.R.; Pathak, R.; Garg, S.; Skinner, C.M.; Melnyk, S.; Pavliv, O.; Hendrickson, H.; Landes, R.D.; Lumen, A.; Tackett, A.J.; et al. Short-term dietary methionine supplementation affects one-carbon metabolism and DNA methylation in the mouse gut and leads to altered microbiome profiles, barrier function, gene expression and histomorphology. Genes Nutr. 2017, 12, 22. [Google Scholar] [CrossRef] [Green Version]
- Quan, L.; Ohgaki, R.; Hara, S.; Okuda, S.; Wei, L.; Okanishi, H.; Nagamori, S.; Endou, H.; Kanai, Y. Amino acid transporter LAT1 in tumor-associated vascular endothelium promotes angiogenesis by regulating cell proliferation and VEGF-A-dependent mTORC1 activation. J. Exp. Clin. Cancer Res. 2020, 39, 266. [Google Scholar] [CrossRef]
- Najumudeen, A.K.; Ceteci, F.; Fey, S.K.; Hamm, G.; Steven, R.T.; Hall, H.; Nikula, C.J.; Dexter, A.; Murta, T.; Race, A.M.; et al. The amino acid transporter SLC7A5 is required for efficient growth of KRAS-mutant colorectal cancer. Nat. Genet. 2021, 53, 16–26. [Google Scholar] [CrossRef]
- Dann, S.G.; Ryskin, M.; Barsotti, A.M.; Golas, J.; Shi, C.; Miranda, M.; Hosselet, C.; Lemon, L.; Lucas, J.; Karnoub, M.; et al. Reciprocal regulation of amino acid import and epigenetic state through Lat1 and EZH2. EMBO J. 2015, 34, 1773–1785. [Google Scholar] [CrossRef] [PubMed]
- Byun, J.K.; Park, M.; Lee, S.; Yun, J.W.; Lee, J.; Kim, J.S.; Cho, S.J.; Jeon, H.J.; Lee, I.K.; Choi, Y.K.; et al. Inhibition of glutamine utilization synergizes with immune checkpoint inhibitor to promote antitumor immunity. Mol. Cell 2020, 80, 592–606. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.; Li, W.; Kremer, D.M.; Sajjakulnukit, P.; Li, S.; Crespo, J.; Nwosu, Z.C.; Zhang, L.; Czerwonka, A.; Pawłowska, A.; et al. Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature 2020, 585, 277–282. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, X.; Yu, Q.; Wang, H.; Tan, F.; Zhu, Q.; Yuan, L.; Jiang, H.; Yu, L.; Zeng, S. Epigenetic activation of the drug transporter OCT2 sensitizes renal cell carcinoma to oxaliplatin. Sci. Transl. Med. 2016, 8, 348ra397. [Google Scholar] [CrossRef] [PubMed]
- Arango, D.; Wilson, A.J.; Shi, Q.; Corner, G.A.; Aranes, M.J.; Nicholas, C.; Lesser, M.; Mariadason, J.M.; Augenlicht, L.H. Molecular mechanisms of action and prediction of response to oxaliplatin in colorectal cancer cells. Br. J. Cancer 2004, 91, 1931–1946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogliolo, S.; Cassani, C.; Gardella, B.; Musacchi, V.; Babilonti, L.; Venturini, P.L.; Ferrero, S.; Spinillo, A. Oxaliplatin for the treatment of ovarian cancer. Expert Opin. Investig. Drugs 2015, 24, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.T.; Lin, Y.T.; Tang, S.P.; Luo, C.K.; Tsai, C.T.; Shun, C.T.; Chen, C.C. Metabolic targeting of HIF-1α potentiates the therapeutic efficacy of oxaliplatin in colorectal cancer. Oncogene 2020, 39, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Browning, R.J.; Reardon, P.J.T.; Parhizkar, M.; Pedley, R.B.; Edirisinghe, M.; Knowles, J.C.; Stride, E. Drug delivery strategies for platinum-based chemotherapy. ACS Nano 2017, 11, 8560–8578. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Chen, L.; Qin, Z.; Lei, J.; Ye, S.; Zeng, K.; Wang, H.; Ying, M.; Gao, J.; Zeng, S.; et al. Upregulation of miR-489-3p and miR-630 inhibits oxaliplatin uptake in renal cell carcinoma by targeting OCT2. Acta Pharm. Sin. B 2019, 9, 1008–1020. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Q.; Liu, Y.; Sun, W.; Song, T.; Jiang, X.; Zeng, K.; Zeng, S.; Chen, L.; Yu, L. Blockade LAT1 Mediates Methionine Metabolism to Overcome Oxaliplatin Resistance under Hypoxia in Renal Cell Carcinoma. Cancers 2022, 14, 2551. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers14102551
Xu Q, Liu Y, Sun W, Song T, Jiang X, Zeng K, Zeng S, Chen L, Yu L. Blockade LAT1 Mediates Methionine Metabolism to Overcome Oxaliplatin Resistance under Hypoxia in Renal Cell Carcinoma. Cancers. 2022; 14(10):2551. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers14102551
Chicago/Turabian StyleXu, Qingwen, Yuxi Liu, Wen Sun, Tiantian Song, Xintong Jiang, Kui Zeng, Su Zeng, Lu Chen, and Lushan Yu. 2022. "Blockade LAT1 Mediates Methionine Metabolism to Overcome Oxaliplatin Resistance under Hypoxia in Renal Cell Carcinoma" Cancers 14, no. 10: 2551. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers14102551