Who’s Who? Discrimination of Human Breast Cancer Cell Lines by Raman and FTIR Microspectroscopy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Sample Preparation for Spectroscopic Analysis
2.3. Raman Microspectroscopy
2.4. FTIR Microspectroscopy
3. Results
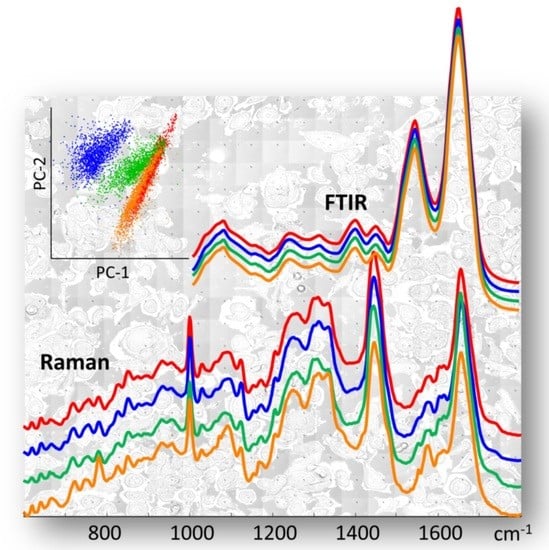
3.1. All Breast Cancer Cells—Overall Characterisation
3.2. TNBC versus Non-TNBC Cells
3.3. TNBC—Mesenchymal Stem-Cell Like versus Basal-like 1 Cells
3.4. TNBC Basal-like 1 Cells—High-Metastatic versus Low-Metastatic Potential
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Li, C.I.; Uribe, D.J.; Daling, J.R. Clinical characteristics of different histologic types of breast cancer. Br. J. Cancer 2005, 93, 1046–1052. [Google Scholar] [CrossRef] [PubMed]
- Lazaro-Pacheco, D.; Shaaban, A.M.; Rehman, S.; Rehman, I. Raman spectroscopy of breast cancer. Appl. Spectrosc. Rev. 2019, 55, 439–475. [Google Scholar] [CrossRef]
- Polyak, K. Heterogeneity in breast cancer. J. Clin. Investig. 2011, 121, 3786–3788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertos, N.R.; Park, M. Breast cancer—One term, many entities? J. Clin. Investig. 2011, 121, 3789–3796. [Google Scholar] [CrossRef]
- Barzaman, K.; Karami, J.; Zarei, Z.; Hosseinzadeh, A.; Kazemi, M.H.; Moradi-Kalbolandi, S.; Safari, E.; Farahmand, L. Breast cancer: Biology, biomarkers, and treatments. Int. Immunopharmacol. 2020, 84, 106535. [Google Scholar] [CrossRef] [PubMed]
- Luond, F.; Tiede, S.; Christofori, G. Breast cancer as an example of tumour heterogeneity and tumour cell plasticity during malignant progression. Br. J. Cancer 2021, 125, 164–175. [Google Scholar] [CrossRef]
- Liu, M.C.; Pitcher, B.N.; Mardis, E.R.; Davies, S.R.; Friedman, P.N.; Snider, J.E.; Vickery, T.L.; Reed, J.P.; DeSchryver, K.; Singh, B.; et al. PAM50 gene signatures and breast cancer prognosis with adjuvant anthracycline- and taxane-based chemotherapy: Correlative analysis of C9741 (Alliance). NPJ Breast Cancer 2016, 2, 1–8. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, B.D.; Jovanovic, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hubalek, M.; Czech, T.; Muller, H. Biological Subtypes of Triple-Negative Breast Cancer. Breast Care 2017, 12, 8–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fougner, C.; Bergholtz, H.; Norum, J.H.; Sorlie, T. Re-definition of claudin-low as a breast cancer phenotype. Nat. Commun. 2020, 11, 1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kassam, F.; Enright, K.; Dent, R.; Dranitsaris, G.; Myers, J.; Flynn, C.; Fralick, M.; Kumar, R.; Clemons, M. Survival outcomes for patients with metastatic triple-negative breast cancer: Implications for clinical practice and trial design. Clin. Breast Cancer 2009, 9, 29–33. [Google Scholar] [CrossRef]
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar] [CrossRef] [Green Version]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-Negative Breast Cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef] [Green Version]
- Bento, M.J.; Goncalves, G.; Aguiar, A.; Castro, C.; Veloso, V.; Rodrigues, V. Performance indicators evaluation of the population-based breast cancer screening programme in Northern Portugal using the European Guidelines. Cancer Epidemiol. 2015, 39, 783–789. [Google Scholar] [CrossRef]
- Zolota, V.; Tzelepi, V.; Piperigkou, Z.; Kourea, H.; Papakonstantinou, E.; Argentou, M.-I.; Karamanos, N.K. Epigenetic Alterations in Triple-Negative Breast Cancer-The Critical Role of Extracellular Matrix. Cancers 2021, 13, 713. [Google Scholar] [CrossRef] [PubMed]
- Plodinec, M.; Loparic, M.; Monnier, C.A.; Obermann, E.C.; Zanetti-Dallenbach, R.; Oertle, P.; Hyotyla, J.T.; Aebi, U.; Bentires-Alj, M.; Lim, R.Y.; et al. The nanomechanical signature of breast cancer. Nat. Nanotechnol. 2012, 7, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Mezencev, R.; Kim, B.; Wang, L.; McDonald, J.; Sulchek, T. Cell stiffness is a biomarker of the metastatic potential of ovarian cancer cells. PLoS ONE 2012, 7, 46609. [Google Scholar] [CrossRef] [Green Version]
- Lekka, M. Discrimination between Normal and Cancerous Cells Using AFM. Bionanoscience 2016, 6, 65–80. [Google Scholar] [CrossRef] [Green Version]
- Marques, M.P.M.; Batista de Carvalho, A.L.M.; Mamede, A.P.; Dopplapudi, A.; Rudic, S.; Tyagi, M.; Garcia Sakai, V.; Batista de Carvalho, L.A.E. A New Look into the Mode of Action of Metal-Based Anticancer Drugs. Molecules 2020, 25, 246. [Google Scholar] [CrossRef] [Green Version]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [Green Version]
- Grosserueschkamp, F.; Kallenbach-Thieltges, A.; Behrens, T.; Brüning, T.; Altmayer, M.; Stamatis, G.; Theegarten, D.; Gerwert, K. Marker-free automated histopathological annotation of lung tumour subtypes by FTIR imaging. Analyst 2015, 140, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Haka, A.S.; Shafer-Peltier, K.E.; Fitzmaurice, M.; Crowe, J.; Dasari, R.R.; Feld, M.S. Diagnosing breast cancer by using Raman spectroscopy. Proc. Natl. Acad. Sci. USA 2005, 102, 12371–12376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medeiros, P.S.; Batista de Carvalho, A.L.M.; Ruano, C.; Otero, J.C.; Marques, M.P. Raman microspectroscopy for probing the impact of a dietary antioxidant on human breast cancer cells. Food Funct. 2016, 7, 2800–2810. [Google Scholar] [CrossRef] [Green Version]
- Butler, H.J.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; White, K.E.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L.; et al. Using Raman spectroscopy to characterise biological materials. Nat. Protoc. 2016, 11, 664–687. [Google Scholar] [CrossRef] [Green Version]
- Hughes, C.; Baker, M.J. Can mid-infrared biomedical spectroscopy of cells, fluids and tissue aid improvements in cancer survival? A patient paradigm. Analyst 2016, 141, 467–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilling, M.; Gardner, P. Fundamental developments in infrared spectroscopic imaging for biomedical applications. Chem. Soc. Rev. 2016, 45, 1935–1957. [Google Scholar] [CrossRef]
- Batista de Carvalho, A.L.M.; Pilling, M.; Gardner, P.; Doherty, J.; Cinque, G.; Wehbe, K.; Kelley, C.; Batista de Carvalho, L.A.E.; Marques, M.P. Chemotherapeutic response to cisplatin-like drugs in human breast cancer cells probed by vibrational microspectroscopy. Faraday Discuss. 2016, 187, 273–298. [Google Scholar] [CrossRef] [Green Version]
- Santos, I.P.; Barroso, E.M.; Bakker Schut, T.C.; Caspers, P.J.; van Lanschot, C.G.F.; Choi, D.-H.; van der Kamp, M.F.; Smits, R.W.H.; van Doorn, R.; Verdijk, R.M.; et al. Raman spectroscopy for cancer detection and cancer surgery guidance: Translation to the clinics. Analyst 2017, 142, 3025–3047. [Google Scholar] [CrossRef]
- Mohamed, H.T.; Untereiner, V.; Proult, I.; Ibrahim, S.A.; Götte, M.; El-Shinawi, M.; Mohamed, M.M.; Sockalingum, G.D.; Brezillon, S. Characterization of inflammatory breast cancer: A vibrational microspectroscopy and imaging approach at the cellular and tissue level. Analyst 2018, 143, 6103–6112. [Google Scholar] [CrossRef] [Green Version]
- Schie, I.W.; Rüger, J.; Mondol, A.S.; Ramoji, A.; Neugebauer, U.; Krafft, C.; Popp, J. High-Throughput Screening Raman Spectroscopy Platform for Label-Free Cellomics. Anal. Chem. 2018, 90, 2023–2030. [Google Scholar] [CrossRef] [PubMed]
- Marques, M.P.M.; Batista de Carvalho, A.L.M.; Mamede, A.P.; Santos, I.P.; Garcia Sakai, V.; Dopplapudi, A.; Cinque, G.; Wolna, M.; Gardner, P.; Batista de Carvalho, L.A.E. Chemotherapeutic Targets in Osteosarcoma: Insights from Synchrotron-MicroFTIR and Quasi-Elastic Neutron Scattering. J. Phys. Chem. B 2019, 123, 6968–6979. [Google Scholar] [CrossRef]
- Elumalai, S.; Manago, S.; De Luca, A.C. Raman Microscopy: Progress in Research on Cancer Cell Sensing. Sensors 2020, 20, 5525. [Google Scholar] [CrossRef]
- Quaroni, L. Infrared microscopy in the study of cellular biochemistry. Infrared Phys. Technol. 2020, 105, 102779. [Google Scholar] [CrossRef]
- Mohamed, H.T.; Untereiner, V.; Cinque, G.; Ibrahim, S.A.; Götte, M.; Nguyen, N.Q.; Rivet, R.; Sockalingum, G.D.; Brezillon, S. Infrared Microspectroscopy and Imaging Analysis of Inflammatory and Non-Inflammatory Breast Cancer Cells and Their GAG Secretome. Molecules 2020, 25, 4300. [Google Scholar] [CrossRef] [PubMed]
- Su, K.Y.; Lee, W.L. Fourier Transform Infrared Spectroscopy as a Cancer Screening and Diagnostic Tool: A Review and Prospects. Cancers 2020, 12, 115. [Google Scholar] [CrossRef] [Green Version]
- Gardner, B.; Matousek, P.; Stone, N. Self-absorption corrected non-invasive transmission Raman spectroscopy (of biological tissue). Analyst 2021, 146, 1260–1267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, C.; Peng, D.; Yi, X.; He, S.; Liu, F.; Zheng, X.; Huang, W.E.; Zhao, L.; Huang, X. Raman spectroscopy and machine learning for the classification of breast cancers. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 264, 120300. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, K.; Araki, A.; Krishna, C.M.; Maruyama, R.; Yamamoto, T.; Noothalapati, H. Identification of Molecular Basis for Objective Discrimination of Breast Cancer Cells (MCF-7) from Normal Human Mammary Epithelial Cells by Raman Microspectroscopy and Multivariate Curve Resolution Analysis. Int. J. Mol. Sci. 2021, 22, 800. [Google Scholar] [CrossRef]
- Zoladek, A.; Pascut, F.C.; Patel, P.; Notingher, I. Non-invasive time-course imaging of apoptotic cells by confocal Raman micro-spectroscopy. J. Raman Spectrosc. 2011, 42, 251–258. [Google Scholar] [CrossRef]
- Wehbe, K.; Filik, J.; Frogley, M.D.; Cinque, G. The effect of optical substrates on micro-FTIR analysis of single mammalian cells. Anal. Bioanal. Chem. 2013, 405, 1311–1324. [Google Scholar] [CrossRef] [Green Version]
- Notingher, I.; Verrier, S.; Haque, S.; Polak, J.M.; Hench, L.L. Spectroscopic Study of Human Lung Epithelial Cells (A549) in Culture: Living Cells Versus Dead Cells. Biopolymers 2002, 72, 230–240. [Google Scholar] [CrossRef]
- Talari, A.C.S.; Evans, C.A.; Holen, I.; Coleman, R.E.; Rehman, I.U. Raman spectroscopic analysis differentiates between breast cancer cell lines. J. Raman Spectrosc. 2015, 46, 421–427. [Google Scholar] [CrossRef]
- Marro, M.; Nieva, C.; Sanz-Pamplona, R.; Sierra, A. Molecular monitoring of epithelial-to-mesenchymal transition in breast cancer cells by means of Raman spectroscopy. Biochim. Biophys. Acta 2014, 1843, 1785–1795. [Google Scholar] [CrossRef] [Green Version]
- Ozek, N.S.; Tuna, S.; Erson-Bensan, A.E.; Severcan, F. Characterization of microRNA-125b expression in MCF7 breast cancer cells by ATR-FTIR spectroscopy. Analyst 2010, 135, 3094–3102. [Google Scholar] [CrossRef]
- Dolce, V.; Rita Cappello, A.; Lappano, R.; Maggiolini, M. Glycerophospholipid synthesis as a novel drug target against cancer. Curr. Mol. Pharmacol. 2011, 4, 167–175. [Google Scholar] [CrossRef]
- Chaturvedi, D.; Balaji, S.A.; Bn, V.K.; Ariese, F.; Umapathy, S.; Rangarajan, A. Different Phases of Breast Cancer Cells: Raman Study of Immortalized, Transformed, and Invasive Cells. Biosensors 2016, 6, 57. [Google Scholar] [CrossRef] [Green Version]
- Abramczyk, H.; Surmacki, J.; Kopec, M.; Olejnik, A.K.; Kaufman-Szymczyk, A.; Fabianowska-Majewska, K. Epigenetic changes in cancer by Raman imaging, fluorescence imaging, AFM and scanning near-field optical microscopy (SNOM). Acetylation in normal and human cancer breast cells MCF10A, MCF7 and MDA-MB-231. Analyst 2016, 141, 5646–5658. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.T.; Scherf, U.; Eisen, M.B.; Perou, C.M.; Rees, C.; Spellman, P.; Iyer, V.; Jeffrey, S.S.; Van de Rijn, M.; Waltham, M.; et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 2000, 24, 227–235. [Google Scholar] [CrossRef]
- Sellappan, S.; Grijalva, R.; Zhou, X.; Yang, W.; Bar Eli, M.; Mills, G.B.; Yu, D. Lineage Infidelity of MDA-MB-435 Cells: Expression of Melanocyte Proteins in a Breast Cancer Cell Line. Cancer Res. 2004, 64, 3479–3485. [Google Scholar] [CrossRef] [Green Version]
- Rae, J.M.; Ramus, S.J.; Waltham, M.; Armes, J.E.; Campbell, I.G.; Clarke, R.; Barndt, R.J.; Johnson, M.D.; Thompson, E.W. Common origins of MDA-MB-435 cells from various sources with those shown to have melanoma properties. Clin. Exp. Metastasis 2004, 21, 543–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rae, J.M.; Creighton, C.J.; Meck, J.M.; Haddad, B.R.; Johnson, M.D. MDA-MB-435 cells are derived from M14 melanoma cells--a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res. Treat. 2007, 104, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Kenny, P.A.; Lee, G.Y.; Myers, C.A.; Neve, R.M.; Semeiks, J.R.; Spellman, P.T.; Lorenz, K.; Lee, E.H.; Barcellos-Hoff, M.H.; Petersen, O.W.; et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 2007, 1, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Ziperstein, M.J.; Guzman, A.; Kaufman, L.J. Breast Cancer Cell Line Aggregate Morphology Does Not Predict Invasive Capacity. PLoS ONE 2015, 10, 0139523. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Bentires-Alj, M. Breast Tumor Heterogeneity: Source of Fitness, Hurdle for Therapy. Mol. Cell 2015, 60, 537–546. [Google Scholar] [CrossRef]
- Prat, A.; Adamo, B.; Cheang, M.C.; Anders, C.K.; Carey, L.A.; Perou, C.M. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist 2013, 18, 123–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Chan, T.; Mak, M. Morphodynamic signatures of MDA-MB-231 single cells and cell doublets undergoing invasion in confined microenvironments. Sci. Rep. 2021, 11, 6529. [Google Scholar] [CrossRef] [PubMed]






| Band (cm−1) | Assignment a | |||
|---|---|---|---|---|
| Nucleic Acids | Proteins | Lipids | Carbohydrates | |
| 640–642 | ν(CS), τ(CC)Tyr | |||
| 670 | (ν(CC)ring)B-DNA/G, T | |||
| 698 | (ν(CC)ring)B-DNA/dG | ν(CS)Met | ||
| 718–722 | (νs(CC)ring)B-DNA/A | (νs(CC)ring)Trp | ||
| 754–756 | (ν(CC)ring)B-DNA/dT ν(OPO)DNA | (νs(CC)ring)Trp | ||
| 780–800 | (ν(CC)ring)C, T, U | |||
| 826–830 | (ν(OPO)backbone)B-DNA | ν(CC)Pro, Tyr | ||
| 850–852 | ν(CC)Pro, Tyr, Val, δ(CCH) | ϒ(COC)polysaccharides | ||
| 874–878 | (ribose, ν(CC)ring)RNA | (ν(CC)ring)Tyr, (ν(CC))Hyp | νs(CCN+)phosphocholine | ν(CC), ν(C-O)ring |
| 890–898 | ρ(CH2) | |||
| 934–936 | ν(CC)(α-helix) | ν(COC)glycolipids | ν(COC)glycosidic | |
| 1000 | (νs(CC)ring)Phe | |||
| 1030 | δ(CH)Phe, ν(O-CH3) | ν(CC), ρ(CH2)phospholipids | ν(CC), ν(CO), ν(C-OH) | |
| 1062 | ν(CO)B-DNA/deoxyribose | ν(CC), ν(CN) | ν(CC), ν(CO) | ν(CC), ν(CO), δ(OCH) |
| 1076–1079 | ν(CC), ν(CN) | νs(PO2−)phospholipids | ν(CC)glycogen, ν(CO) | |
| 1092 | νs(PO2−)B-DNA | |||
| 1124 | ν(CO)RNA/ribose | ν(CN) | ν(CC)acyl (trans conformation) | ν(CO), ν(CC) |
| 1157 | ν(CC), ν(CN), ρCH2) | ρ(CH2), ν(=C-C=)conjugated | ρ(CH2) | |
| 1172 | (ν(CC)ring)C,G,T | δ(CH)Tyr, Phe | ||
| 1206 | ν(CC)Hyp, Phe, Tyr | |||
| 1234 | amide III/β-sheet | |||
| 1238 | (νas(PO2−))B-DNA | |||
| 1250 | amide III/ random coil | |||
| 1252–1264 | (ν(CC)ring)A,T | ω(CH2), δ(C=C-H) | ω(CH2),δ(C=C-H)phospholipids | ω(CH2), t(CH2) |
| 1272 | amide III/α-helix | ω(CH2), t(CH2) | ω(CH2), t(CH2) | |
| 1302 | (ν(CC)ring)RNA/A,C | |||
| 1312 | (ν(CC)ring)G | t(CH2) | t(CH2) | t(CH2) |
| 1334 | (ν(CC)ring)G | |||
| 1372 b | (ν(CC)ring)A,G,T | (δs(CH3))glycoproteins | (δs(CH3))lipids/acyl chains | (ω(CH2))saccharides |
| 1400 | t(NH2) | |||
| 1444–1450 | α(CH2) | |||
| 1480 b | δs(NH3+) | |||
| 1508 | Tyr (ν(CC)ring) | |||
| 1546 | ((δ(CN-H)/ν(CN))amide II | |||
| 1552 | (ν(CC)ring)G | (ν(CC)ring)Trp, ν(C=C)porphyrin | ||
| 1572–1574 | (ν(CC)ring)A,G | |||
| 1580 | ν(C=C), ν(C=N) | ν(C=C), ν(C=N) | ||
| 1602 | (ν(CC)ring)T | (δ(C=CH))Phe | ||
| 1612–1690 | amide I/β-sheet antiparallel | |||
| 1614 | (ν(CC)ring)A, δ(NH2)C | ν(C=C)Phe, Tyr, Trp, α(NH2) | ||
| 1612–1690 | amide I/β-sheet antiparallel | |||
| 1627–1640 | amide I/β-sheet antiparallel | |||
| 1640–1650 | amide I/random coil | |||
| 1650 | δ(NH)DNA | ν(C=O)amide I/α-helix | ν(C=C) | |
| 1690 | ν(C=O)amino acid side chain | |||
| 1717 | ν(C=O)B-DNA | |||
| 1730–42 | (ν(C=O)ester)phospholipids | |||
| 2846–2854 | νs(CH2) | νs(CH2) | νs(CH2) | |
| 2873–2878 | νs(CH3) | νs(CH3) | νs(CH3) | |
| 2892 | νas(CH2) | νas(CH2) | νas(CH2) | |
| 2919 | νas(CH2) | νas(CH2) | νas(CH2) | |
| 2924–2930 | νas(CH2) | νas(CH2) | νas(CH2) | |
| 2954–2958 | νas(CH3) | |||
| 2964 | νas(CH3) | νas(CH3) | νas(CH3) | |
| 3070 | amide B | |||
| 3250–3290 | ν(NH)amide A | |||
| 3340–3370 | ν(OH) | |||
| Classified as | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| MCF-7 | MDA-MB-231 | HCC-1143 | MDA-MB-468 | ROC AUC | Sensitivity (%) | Specificity (%) | Accuracy (%) | ||
| True class | MCF-7 | 415 | 0 | 11 | 22 | 0.97 | 92.6 | 90.9 | 91.4 |
| MDA-MB-231 | 0 | 450 | 1 | 0 | 0.99 | 99.7 | 100.0 | 99.9 | |
| HCC-1143 | 31 | 0 | 347 | 15 | 0.97 | 88.3 | 97.6 | 95.2 | |
| MDA-MB-468 | 71 | 0 | 17 | 192 | 0.96 | 68.6 | 97.1 | 92.0 | |
| Classified as | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| MCF-7 | MDA-MB-231 | HCC-1143 | MDA-MB-468 | ROC AUC | Sensitivity (%) | Specificity (%) | Accuracy (%) | ||
| True class | MCF-7 | 63 | 15 | 5 | 4 | 0.90 | 72.2 | 89.8 | 85.8 |
| MDA-MB-231 | 25 | 48 | 1 | 2 | 0.89 | 63.6 | 93.8 | 87.6 | |
| HCC-1143 | 3 | 1 | 83 | 1 | 0.97 | 94.3 | 96.2 | 95.8 | |
| MDA-MB-468 | 2 | 3 | 5 | 119 | 0.97 | 92.2 | 97.2 | 95.5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, I.P.; Martins, C.B.; Batista de Carvalho, L.A.E.; Marques, M.P.M.; Batista de Carvalho, A.L.M. Who’s Who? Discrimination of Human Breast Cancer Cell Lines by Raman and FTIR Microspectroscopy. Cancers 2022, 14, 452. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers14020452
Santos IP, Martins CB, Batista de Carvalho LAE, Marques MPM, Batista de Carvalho ALM. Who’s Who? Discrimination of Human Breast Cancer Cell Lines by Raman and FTIR Microspectroscopy. Cancers. 2022; 14(2):452. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers14020452
Chicago/Turabian StyleSantos, Inês P., Clara B. Martins, Luís A. E. Batista de Carvalho, Maria P. M. Marques, and Ana L. M. Batista de Carvalho. 2022. "Who’s Who? Discrimination of Human Breast Cancer Cell Lines by Raman and FTIR Microspectroscopy" Cancers 14, no. 2: 452. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers14020452