Automated Machine Learning for Healthcare and Clinical Notes Analysis
Abstract
:1. Introduction
- Towards AutoML for clinical notes analysis: The papers in this category cover ML research to extract diagnoses from clinical notes. We discuss how previous ML methods used for medical notes analysis can be used towards AutoML for clinical notes diagnoses.
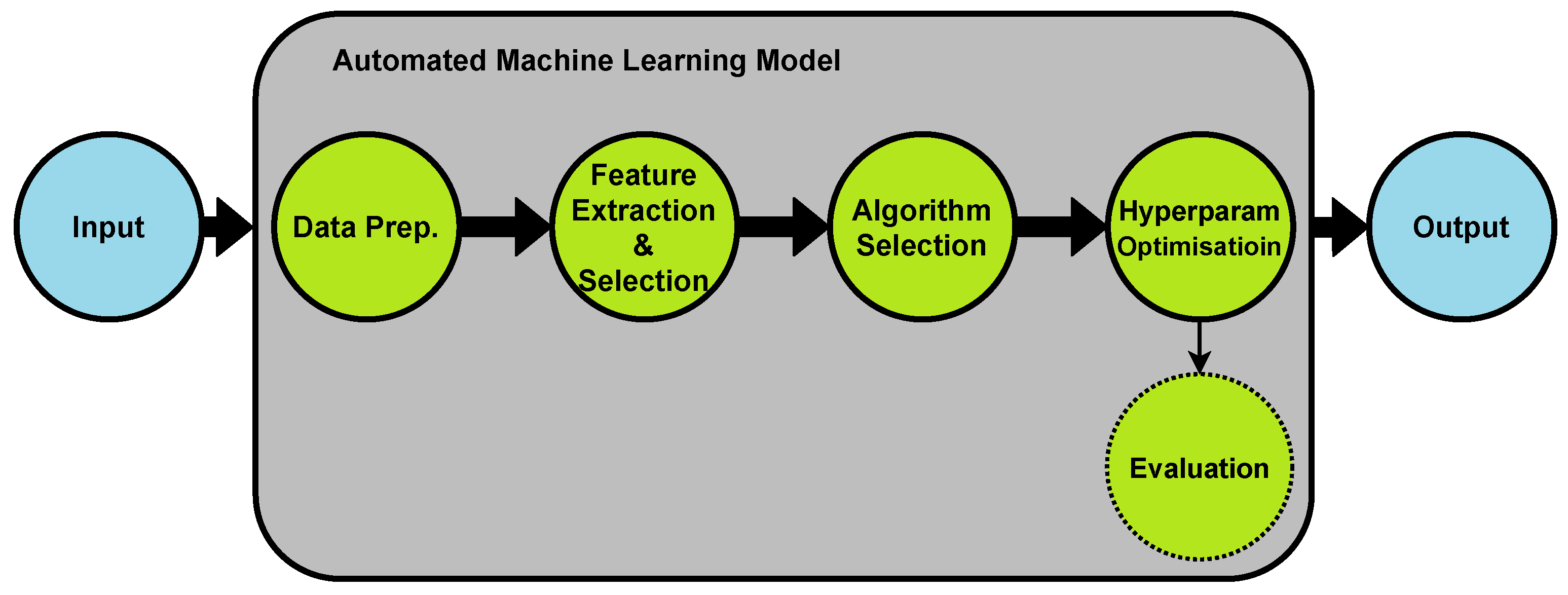
2. Automated Machine Learning (AutoML)
3. AutoML in Healthcare Industry
3.1. Building AutoML for Clinical Datasets
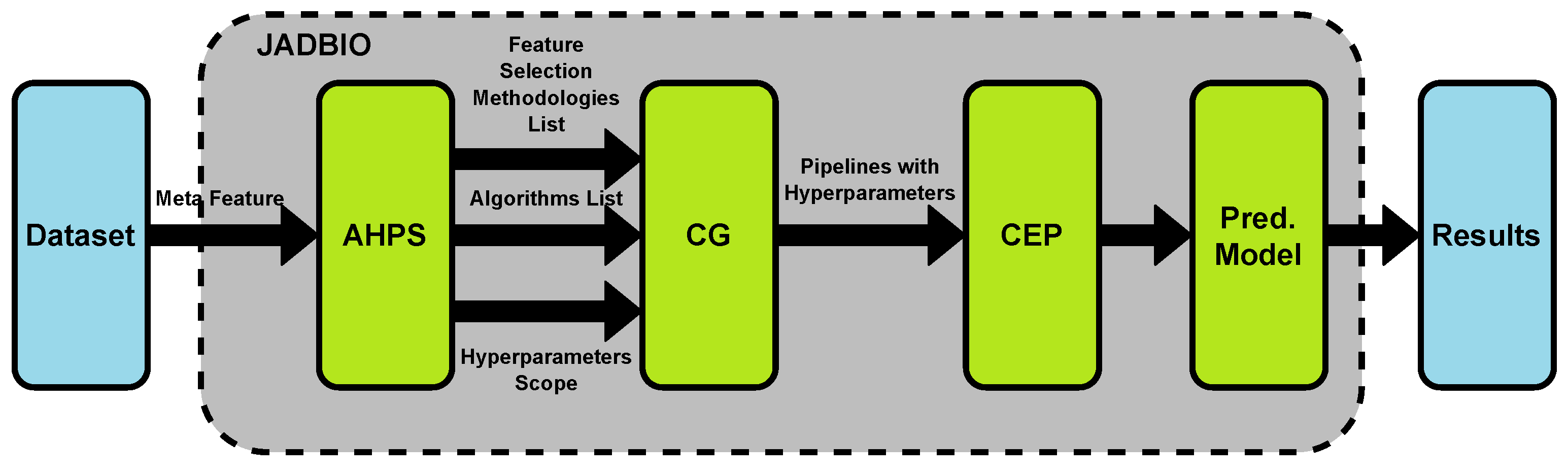
3.2. Using Existing AutoML Tools for Clinical Datasets
- Google AutoML Vision would be the easiest tool to work with medical images. It does not require tool installation or coding. Clinical practitioner can upload image datasets to train their model and then use it for diagnoses. Previous examples include cancer [12] and pneumonia [63] detection based on X-ray images.
- For assessing patients risks using biometrics and patient’s medical history, AutoPrognosis can be used. Use case examples include prognosis of cardiovascular disease [25].
4. Towards AutoML for Clinical Notes
4.1. Challenges in Working with Clinical Notes
4.2. Benefits of Developing AutoML for Clinical Notes
4.3. Machine Learning for Clinical Notes Analysis
4.3.1. Preprocessing
Preprocessing in AutoML for Clinical Notes
4.3.2. Feature Extraction
Feature Extraction in AutoML for Clinical Notes
- Which feature-extraction method or combination of methods should be used in an AutoML for clinical notes?
- Which methods should be used to compare and decide the best feature-extraction techniques?
- Can cTAKES be easily integrated in an AutoML tool for clinical notes analysis? What are the hurdles?
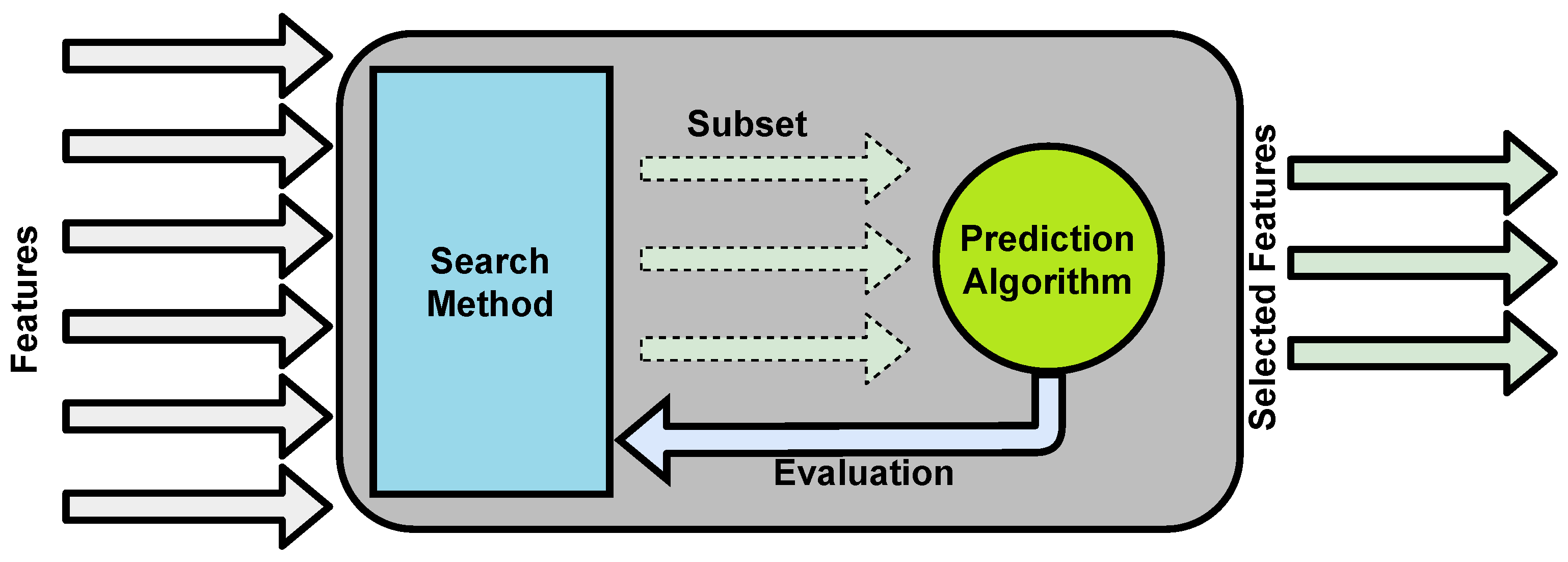
4.3.3. Feature Selection
Feature Selection in AutoML for Clinical Notes
4.3.4. Algorithms Selection and Optimization
Algorithm Selection and Hyperparameter Optimization in AutoML for Clinical Notes
4.3.5. Targets and Evaluations
Evaluation Metrics in AutoML for Clinical Notes
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Azghadi, M.R.; Lammie, C.; Eshraghian, J.K.; Payvand, M.; Donati, E.; Linares-Barranco, B.; Indiveri, G. Hardware Implementation of Deep Network Accelerators Towards Healthcare and Biomedical Applications. IEEE Trans. Biomed. Circuits Syst. 2020, 14, 1138–1159. [Google Scholar] [CrossRef] [PubMed]
- Rong, G.; Mendez, A.; Assi, E.B.; Zhao, B.; Sawan, M. Artificial Intelligence in Healthcare: Review and Prediction Case Studies. Engineering 2020, 6, 291–301. [Google Scholar] [CrossRef]
- Beam, A.L.; Kohane, I.S. Big data and machine learning in health care. JAMA 2018, 319, 1317–1318. [Google Scholar] [CrossRef]
- Li, J.P.; Haq, A.U.; Din, S.U.; Khan, J.; Khan, A.; Saboor, A. Heart Disease Identification Method Using Machine Learning Classification in E-Healthcare. IEEE Access 2020, 8, 107562–107582. [Google Scholar] [CrossRef]
- Leite, A.F.; Vasconcelos, K.d.F.; Willems, H.; Jacobs, R. Radiomics and machine learning in oral healthcare. Proteom. Clin. Appl. 2020, 14, 1900040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- Feurer, M.; Klein, A.; Eggensperger, K.; Springenberg, J.; Blum, M.; Hutter, F. Efficient and robust automated machine learning. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; pp. 2962–2970. [Google Scholar]
- Hutter, F.; Kotthoff, L.; Vanschoren, J. Automated Machine Learning: Methods, Systems, Challenges; Springer: Cham, Switzerland, 2019. [Google Scholar]
- Yao, Q.; Wang, M.; Chen, Y.; Dai, W.; Li, Y.F.; Tu, W.W.; Yang, Q.; Yu, Y. Taking human out of learning applications: A survey on automated machine learning. arXiv 2018, arXiv:1810.13306. [Google Scholar]
- Waring, J.; Lindvall, C.; Umeton, R. Automated machine learning: Review of the state-of-the-art and opportunities for healthcare. Artif. Intell. Med. 2020, 104, 101822. [Google Scholar] [CrossRef]
- Ooms, R.; Spruit, M. Self-Service Data Science in Healthcare with Automated Machine Learning. Appl. Sci. 2020, 10, 2992. [Google Scholar] [CrossRef]
- Borkowski, A.A.; Wilson, C.P.; Borkowski, S.A.; Thomas, L.B.; Deland, L.A.; Grewe, S.J.; Mastorides, S.M. Google Auto ML versus Apple Create ML for Histopathologic Cancer Diagnosis; Which Algorithms Are Better? arXiv 2019, arXiv:1903.08057. [Google Scholar]
- Tsamardinos, I.; Charonyktakis, P.; Lakiotaki, K.; Borboudakis, G.; Zenklusen, J.C.; Juhl, H.; Chatzaki, E.; Lagani, V. Just Add Data: Automated Predictive Modeling and BioSignature Discovery. bioRxiv 2020. [Google Scholar] [CrossRef]
- Karaglani, M.; Gourlia, K.; Tsamardinos, I.; Chatzaki, E. Accurate Blood-Based Diagnostic Biosignatures for Alzheimer’s Disease via Automated Machine Learning. J. Clin. Med. 2020, 9, 3016. [Google Scholar] [CrossRef] [PubMed]
- Gehrmann, S.; Dernoncourt, F.; Li, Y.; Carlson, E.T.; Wu, J.T.; Welt, J.; Foote, J., Jr.; Moseley, E.T.; Grant, D.W.; Tyler, P.D. Comparing rule-based and deep learning models for patient phenotyping. arXiv 2017, arXiv:1703.08705. [Google Scholar]
- Nigam, P. Applying Deep Learning to ICD-9 Multi-Label Classification from Medical Records; Technical Report; Stanford University: Stanford, CA, USA, 2016. [Google Scholar]
- Venkataraman, G.R.; Pineda, A.L.; Bear Don’t Walk IV, O.J.; Zehnder, A.M.; Ayyar, S.; Page, R.L.; Bustamante, C.D.; Rivas, M.A. FasTag: Automatic text classification of unstructured medical narratives. PLoS ONE 2020, 15, e0234647. [Google Scholar] [CrossRef] [PubMed]
- Yogarajan, V.; Montiel, J.; Smith, T.; Pfahringer, B. Seeing The Whole Patient: Using Multi-Label Medical Text Classification Techniques to Enhance Predictions of Medical Codes. arXiv 2020, arXiv:2004.00430. [Google Scholar]
- Boytcheva, S. Automatic matching of ICD-10 codes to diagnoses in discharge letters. In Proceedings of the Second Workshop on Biomedical Natural Language Processing, Hissar, Bulgaria, 15 September 2011; pp. 11–18. [Google Scholar]
- Huang, J.; Osorio, C.; Sy, L.W. An empirical evaluation of deep learning for ICD-9 code assignment using MIMIC-III clinical notes. Comput. Methods Programs Biomed. 2019, 177, 141–153. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Chapman, W.W.; Miller, T.A.; Lin, C.; Crowley, R.S.; Savova, G.K. A system for coreference resolution for the clinical narrative. J. Am. Med. Inform. Assoc. 2012, 19, 660–667. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Wagholikar, K.B.; Jonnalagadda, S.; Sohn, S. Integrated cTAKES for Concept Mention Detection and Normalization. In Proceedings of the 2013 Cross Language Evaluation Forum Conference, Valencia, Spain, 23–26 September 2013. [Google Scholar]
- Mullenbach, J.; Wiegreffe, S.; Duke, J.; Sun, J.; Eisenstein, J. Explainable prediction of medical codes from clinical text. arXiv 2018, arXiv:1802.05695. [Google Scholar]
- Bisong, E. Google AutoML: Cloud Vision. In Building Machine Learning and Deep Learning Models on Google Cloud Platform; Springer: Berlin/Heidelberg, Germany, 2019; pp. 581–598. [Google Scholar]
- Alaa, A.M.; van der Schaar, M. Autoprognosis: Automated clinical prognostic modeling via bayesian optimization with structured kernel learning. arXiv 2018, arXiv:1802.07207. [Google Scholar]
- Baškarada, S.; Koronios, A. Unicorn data scientist: The rarest of breeds. Program 2017, 51, 65–74. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, C.; Yang, Q. Data preparation for data mining. Appl. Artif. Intell. 2003, 17, 375–381. [Google Scholar] [CrossRef]
- Khalid, S.; Khalil, T.; Nasreen, S. A survey of feature selection and feature extraction techniques in machine learning. In Proceedings of the 2014 Science and Information Conference, London, UK, 27–29 August 2014; pp. 372–378. [Google Scholar]
- Yang, C.; Akimoto, Y.; Kim, D.W.; Udell, M. OBOE: Collaborative filtering for AutoML model selection. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage, AK, USA, 4–8 August 2019; pp. 1173–1183. [Google Scholar]
- Shahriari, B.; Swersky, K.; Wang, Z.; Adams, R.P.; De Freitas, N. Taking the human out of the loop: A review of Bayesian optimization. Proc. IEEE 2015, 104, 148–175. [Google Scholar] [CrossRef] [Green Version]
- Thornton, C.; Hutter, F.; Hoos, H.H.; Leyton-Brown, K. Auto-WEKA: Combined selection and hyperparameter optimization of classification algorithms. In Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Chicago, IL, USA, 11–14 August 2013; pp. 847–855. [Google Scholar]
- Kotthoff, L.; Thornton, C.; Hoos, H.H.; Hutter, F.; Leyton-Brown, K. Auto-WEKA 2.0: Automatic model selection and hyperparameter optimization in WEKA. J. Mach. Learn. Res. 2017, 18, 826–830. [Google Scholar]
- Jungermann, F. Information Extraction with Rapidminer. Available online: https://duepublico2.uni-due.de/servlets/MCRFileNodeServlet/duepublico_derivate_00038023/Tagungsband_GSCLSYMP2009_final_6.pdf (accessed on 20 February 2021).
- Gosiewska, A.; Bakala, M.; Woznica, K.; Zwolinski, M.; Biecek, P. EPP: Interpretable score of model predictive power. arXiv 2019, arXiv:1908.09213. [Google Scholar]
- Perotte, A.; Pivovarov, R.; Natarajan, K.; Weiskopf, N.; Wood, F.; Elhadad, N. Diagnosis code assignment: Models and evaluation metrics. J. Am. Med. Inf. Assoc. 2014, 21, 231–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, J.; Magoulas, R. 2015 Data Science Salary Survey; O’Reilly Media, Incorporated: Sebastopol, CA, USA, 2015. [Google Scholar]
- Luo, G.; Stone, B.L.; Johnson, M.D.; Tarczy-Hornoch, P.; Wilcox, A.B.; Mooney, S.D.; Sheng, X.; Haug, P.J.; Nkoy, F.L. Automating construction of machine learning models with clinical big data: Proposal rationale and methods. JMIR Res. Protoc. 2017, 6, e175. [Google Scholar] [CrossRef] [PubMed]
- Baars, H.; Kemper, H.G. Management support with structured and unstructured data—An integrated business intelligence framework. Inf. Syst. Manag. 2008, 25, 132–148. [Google Scholar] [CrossRef]
- Zhang, D.; Yin, C.; Zeng, J.; Yuan, X.; Zhang, P. Combining structured and unstructured data for predictive models: A deep learning approach. BMC Med. Inform. Decis. Mak. 2020, 20, 1–11. [Google Scholar] [CrossRef]
- Miiro, F.; Nääs, M. SQL and NoSQL Databases: A Case Study in the Azure Cloud. Bachelor’ s Thesis, KTH Royal Institute of Technology, Stockholm, Sweden, 2015. [Google Scholar]
- Barrenechea, M.J.; Jenkins, T. Enterprise Information Management: The Next Generation of Enterprise Software; OpenText: Waterloo, ON, Canada, 2013. [Google Scholar]
- Luo, G. A review of automatic selection methods for machine learning algorithms and hyper-parameter values. Netw. Model. Anal. Health Inform. Bioinform. 2016, 5, 18. [Google Scholar] [CrossRef]
- Zhang, Y.; Bahadori, M.T.; Su, H.; Sun, J. FLASH: Fast Bayesian optimization for data analytic pipelines. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 13–16 August 2016; pp. 2065–2074. [Google Scholar]
- Kim, S.; Kim, I.; Lim, S.; Baek, W.; Kim, C.; Cho, H.; Yoon, B.; Kim, T. Scalable neural architecture search for 3d medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2019; pp. 220–228. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Weng, Y.; Zhou, T.; Li, Y.; Qiu, X. Nas-unet: Neural architecture search for medical image segmentation. IEEE Access 2019, 7, 44247–44257. [Google Scholar] [CrossRef]
- Olson, R.S.; Moore, J.H. TPOT: A tree-based pipeline optimization tool for automating machine learning. Proc. Mach. Learn. Res. 2016, 64, 66–74. [Google Scholar]
- Jin, H.; Song, Q.; Hu, X. Auto-keras: An efficient neural architecture search system. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage, AK, USA, 4–8 August 2019; pp. 1946–1956. [Google Scholar]
- Drori, I.; Krishnamurthy, Y.; Rampin, R.; Lourenço, R.; One, J.; Cho, K.; Silva, C.; Freire, J. AlphaD3M: Machine learning pipeline synthesis. In Proceedings of the AutoML Workshop at ICML, Stockholm, Sweden, 14 July 2018. [Google Scholar]
- Mendoza, H.; Klein, A.; Feurer, M.; Springenberg, J.T.; Hutter, F. Towards automatically-tuned neural networks. In Proceedings of the Workshop on Automatic Machine Learning, New York, NY, USA, 24 June 2016; pp. 58–65. [Google Scholar]
- Swearingen, T.; Drevo, W.; Cyphers, B.; Cuesta-Infante, A.; Ross, A.; Veeramachaneni, K. ATM: A distributed, collaborative, scalable system for automated machine learning. In Proceedings of the 2017 IEEE International Conference on Big Data (Big Data), Boston, MA, USA, 11–14 December 2017; pp. 151–162. [Google Scholar]
- Komer, B.; Bergstra, J.; Eliasmith, C. Hyperopt-sklearn: Automatic hyperparameter configuration for scikit-learn. In Proceedings of the Scientific Computing with Python, Austin, TX, USA, 6–12 July 2014. [Google Scholar]
- Mohr, F.; Wever, M.; Hüllermeier, E. ML-Plan: Automated machine learning via hierarchical planning. Mach. Learn. 2018, 107, 1495–1515. [Google Scholar] [CrossRef] [Green Version]
- Feurer, M.; Eggensperger, K.; Falkner, S.; Lindauer, M.; Hutter, F. Practical automated machine learning for the automl challenge 2018. In Proceedings of the International Workshop on Automatic Machine Learning at ICML, Stockholm, Sweden, 14 July 2018; pp. 1189–1232. [Google Scholar]
- de Sá, A.G.; Pinto, W.J.G.; Oliveira, L.O.V.; Pappa, G.L. RECIPE: A grammar-based framework for automatically evolving classification pipelines. In Proceedings of the European Conference on Genetic Programming; Springer: Berlin/Heidelberg, Germany, 2017; pp. 246–261. [Google Scholar]
- Gijsbers, P.; Vanschoren, J.; Olson, R.S. Layered TPOT: Speeding up tree-based pipeline optimization. arXiv 2018, arXiv:1801.06007. [Google Scholar]
- Chen, B.; Wu, H.; Mo, W.; Chattopadhyay, I.; Lipson, H. Autostacker: A compositional evolutionary learning system. In Proceedings of the Genetic and Evolutionary Computation Conference, Kyoto, Japan, 15–19 July 2018; pp. 402–409. [Google Scholar]
- Dafflon, J.; Pinaya, W.H.; Turkheimer, F.; Cole, J.H.; Leech, R.; Harris, M.A.; Cox, S.R.; Whalley, H.C.; McIntosh, A.M.; Hellyer, P.J. An automated machine learning approach to predict brain age from cortical anatomical measures. Hum. Brain Mapp. 2020. [Google Scholar] [CrossRef]
- Su, X.; Chen, N.; Sun, H.; Liu, Y.; Yang, X.; Wang, W.; Zhang, S.; Tan, Q.; Su, J.; Gong, Q. Automated machine learning based on radiomics features predicts H3 K27M mutation in midline gliomas of the brain. Neuro-oncology 2020, 22, 393–401. [Google Scholar] [CrossRef]
- Orlenko, A.; Moore, J.H.; Orzechowski, P.; Olson, R.S.; Cairns, J.; Caraballo, P.J.; Weinshilboum, R.M.; Wang, L.; Breitenstein, M.K. Considerations for Automated Machine Learning in Clinical Metabolic Profiling: Altered Homocysteine Plasma Concentration Associated with Metformin Exposure. Biocomputing 2018, 23, 460–471. [Google Scholar]
- Zeng, Y.; Zhang, J. A machine learning model for detecting invasive ductal carcinoma with Google Cloud AutoML Vision. Comput. Biol. Med. 2020, 122, 103861. [Google Scholar] [CrossRef]
- Mantas, J. Setting up an Easy-to-Use Machine Learning Pipeline for Medical Decision Support: A Case Study for COVID-19 Diagnosis Based on Deep Learning with CT Scans. Importance Health Inform. Public Health Pandemic 2020, 272, 13. [Google Scholar]
- Faes, L.; Wagner, S.K.; Fu, D.J.; Liu, X.; Korot, E.; Ledsam, J.R.; Back, T.; Chopra, R.; Pontikos, N.; Kern, C. Automated deep learning design for medical image classification by health-care professionals with no coding experience: A feasibility study. Lancet Digit. Health 2019, 1, e232–e242. [Google Scholar] [CrossRef] [Green Version]
- Puri, M. Automated machine learning diagnostic support system as a computational biomarker for detecting drug-induced liver injury patterns in whole slide liver pathology images. Assay Drug Dev. Technol. 2020, 18, 1–10. [Google Scholar] [CrossRef]
- Kim, I.K.; Lee, K.; Park, J.H.; Baek, J.; Lee, W.K. Classification of pachychoroid disease on ultrawide-field indocyanine green angiography using auto-machine learning platform. Br. J. Ophthalmol. 2020. [Google Scholar] [CrossRef]
- Kocbek, S.; Kocbek, P.; Zupanic, T.; Stiglic, G.; Gabrys, B. Using (Automated) Machine Learning and Drug Prescription Records to Predict Mortality and Polypharmacy in Older Type 2 Diabetes Mellitus Patients. In Proceedings of the International Conference on Neural Information Processing; Springer: Berlin/Heidelberg, Germany, 2019; pp. 624–632. [Google Scholar]
- Bhat, G.S.; Shankar, N.; Panahi, I.M. Automated machine learning based speech classification for hearing aid applications and its real-time implementation on smartphone. Annu Int Conf IEEE Eng Med Biol Soc. 2020, 956–959. [Google Scholar] [CrossRef]
- Truong, A.; Walters, A.; Goodsitt, J.; Hines, K.; Bruss, C.B.; Farivar, R. Towards automated machine learning: Evaluation and comparison of automl approaches and tools. In Proceedings of the 2019 IEEE 31st International Conference on Tools with Artificial Intelligence (ICTAI), Portland, OR, USA, 4–6 November 2019; pp. 1471–1479. [Google Scholar]
- Tsanas, A.; Little, M.; McSharry, P.; Ramig, L. Accurate telemonitoring of Parkinson’s disease progression by non-invasive speech tests. Nat. Preced. 2009. [Google Scholar] [CrossRef]
- Khan, W.; Daud, A.; Nasir, J.A.; Amjad, T. A survey on the state-of-the-art machine learning models in the context of NLP. Kuwait J. Sci. 2016, 43, 95–113. [Google Scholar]
- Weng, W.H.; Wagholikar, K.B.; McCray, A.T.; Szolovits, P.; Chueh, H.C. Medical subdomain classification of clinical notes using a machine learning-based natural language processing approach. BMC Med. Inform. Decis. Mak. 2017, 17, 1–13. [Google Scholar] [CrossRef]
- Gupta, S.; MacLean, D.L.; Heer, J.; Manning, C.D. Induced lexico-syntactic patterns improve information extraction from online medical forums. J. Am. Med. Inform. Assoc. 2014, 21, 902–909. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Krishnamurthy, R.; Raghavan, S.; Vaithyanathan, S.; Jagadish, H. Regular expression learning for information extraction. In Proceedings of the 2008 Conference on Empirical Methods in Natural Language Processing, Honolulu, HI, USA, 25–27 October 2008; pp. 21–30. [Google Scholar]
- Wei, Q.; Ji, Z.; Li, Z.; Du, J.; Wang, J.; Xu, J.; Xiang, Y.; Tiryaki, F.; Wu, S.; Zhang, Y. A study of deep learning approaches for medication and adverse drug event extraction from clinical text. J. Am. Med. Inform. Assoc. 2020, 27, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R. A comparative analysis of selected set of natural language processing (NLP) and machine learning (ML) algorithms for clinical coding using clinical classification standards. Stud. Health Technol. Inform. 2018, 252, 73–79. [Google Scholar]
- Cai, T.; Giannopoulos, A.A.; Yu, S.; Kelil, T.; Ripley, B.; Kumamaru, K.K.; Rybicki, F.J.; Mitsouras, D. Natural language processing technologies in radiology research and clinical applications. Radiographics 2016, 36, 176–191. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Hogan, W.R.; Crowley, R.S. Natural language processing methods and systems for biomedical ontology learning. J. Biomed. Inform. 2011, 44, 163–179. [Google Scholar] [CrossRef] [Green Version]
- Medori, J.; Fairon, C. Machine learning and features selection for semi-automatic ICD-9-CM encoding. In Proceedings of the NAACL HLT 2010 Second Louhi Workshop on Text and Data Mining of Health Documents, Los Angeles, CA, USA, 5 June 2010; pp. 84–89. [Google Scholar]
- Pakhomov, S.; Chute, C.G. A Hybrid Approach to Determining Modification of Clinical Diagnoses. In Proceedings of the AMIA Annual Symposium Proceedings, Washington, DC, USA, 11–15 November 2006; American Medical Informatics Association: Bethesda, MD, USA, 2006; p. 609. [Google Scholar]
- Estevez-Velarde, S.; Gutiérrez, Y.; Montoyo, A.; Almeida-Cruz, Y. AutoML strategy based on grammatical evolution: A case study about knowledge discovery from text. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, Florence, Italy, 28 July–2 August 2019; pp. 4356–4365. [Google Scholar]
- Feurer, M.; Eggensperger, K.; Falkner, S.; Lindauer, M.; Hutter, F. Auto-sklearn 2.0: The next generation. arXiv 2020, arXiv:2007.04074. [Google Scholar]
- Wang, Y.; Sohn, S.; Liu, S.; Shen, F.; Wang, L.; Atkinson, E.J.; Amin, S.; Liu, H. A clinical text classification paradigm using weak supervision and deep representation. BMC Med. Inform. Decis. Mak. 2019, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, H.M.; Panahiazar, M.; Liang, A.; Lituiev, D.; Chang, P.; Sohn, J.H.; Chen, Y.Y.; Franc, B.L.; Joe, B.; Hadley, D. Large scale semi-automated labeling of routine free-text clinical records for deep learning. J. Digit. Imaging 2019, 32, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Alzoubi, H.; Ramzan, N.; Alzubi, R.; Mesbahi, E. An Automated System for Identifying Alcohol Use Status from Clinical Text. In Proceedings of the 2018 International Conference on Computing, Electronics & Communications Engineering (iCCECE), Southend, UK, 16–17 August 2018; pp. 41–46. [Google Scholar]
- Xu, K.; Lam, M.; Pang, J.; Gao, X.; Band, C.; Mathur, P.; Papay, F.; Khanna, A.K.; Cywinski, J.B.; Maheshwari, K. Multimodal machine learning for automated ICD coding. In Proceedings of the Machine Learning for Healthcare Conference, PMLR, Ann Arbor, MI, USA, 8–10 August 2019; pp. 197–215. [Google Scholar]
- Aronson, A.R.; Bodenreider, O.; Demner-Fushman, D.; Fung, K.W.; Lee, V.K.; Mork, J.G.; Névéol, A.; Peters, L.; Rogers, W.J. From indexing the biomedical literature to coding clinical text: Experience with MTI and machine learning approaches. In Proceedings of the Biological, Translational, and Clinical Language Processing, Prague, Czech Republic, 29 June 2007; pp. 105–112. [Google Scholar]
- Obeid, J.S.; Weeda, E.R.; Matuskowitz, A.J.; Gagnon, K.; Crawford, T.; Carr, C.M.; Frey, L.J. Automated detection of altered mental status in emergency department clinical notes: A deep learning approach. BMC Med. Inform. Decis. Mak. 2019, 19, 164. [Google Scholar] [CrossRef] [Green Version]
- Soguero-Ruiz, C.; Hindberg, K.; Rojo-Álvarez, J.L.; Skrøvseth, S.O.; Godtliebsen, F.; Mortensen, K.; Revhaug, A.; Lindsetmo, R.O.; Augestad, K.M.; Jenssen, R. Support vector feature selection for early detection of anastomosis leakage from bag-of-words in electronic health records. IEEE J. Biomed. Health Inform. 2014, 20, 1404–1415. [Google Scholar] [CrossRef]
- Atutxa, A.; Pérez, A.; Casillas, A. Machine learning approaches on diagnostic term encoding with the ICD for clinical documentation. IEEE J. Biomed. Health Inform. 2017, 22, 1323–1329. [Google Scholar] [CrossRef]
- Kalyan, K.S.; Sangeetha, S. Secnlp: A survey of embeddings in clinical natural language processing. J. Biomed. Inform. 2020, 101, 103323. [Google Scholar] [CrossRef]
- Shi, H.; Xie, P.; Hu, Z.; Zhang, M.; Xing, E.P. Towards automated ICD coding using deep learning. arXiv 2017, arXiv:1711.04075. [Google Scholar]
- Polignano, M.; Suriano, V.; Lops, P.; de Gemmis, M.; Semeraro, G. A study of Machine Learning models for Clinical Coding of Medical Reports at CodiEsp 2020. In Proceedings of the Working Notes of Conference and Labs of the Evaluation (CLEF) Forum, CEUR Workshop Proceedings, Thessaloniki, Greece, 2–25 September 2020. [Google Scholar]
- Karmakar, A. Classifying medical notes into standard disease codes using Machine Learning. arXiv 2018, arXiv:1802.00382. [Google Scholar]
- Dubois, S.; Romano, N. Learning effective embeddings from medical notes. arXiv 2017, arXiv:1705.07025. [Google Scholar]
- Lin, C.; Hsu, C.J.; Lou, Y.S.; Yeh, S.J.; Lee, C.C.; Su, S.L.; Chen, H.C. Artificial intelligence learning semantics via external resources for classifying diagnosis codes in discharge notes. J. Med. Internet Res. 2017, 19, e380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savova, G.K.; Masanz, J.J.; Ogren, P.V.; Zheng, J.; Sohn, S.; Kipper-Schuler, K.C.; Chute, C.G. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): Architecture, component evaluation and applications. J. Am. Med. Inform. Assoc. 2010, 17, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Torii, M.; Wagholikar, K.; Liu, H. Using machine learning for concept extraction on clinical documents from multiple data sources. J. Am. Med. Inform. Assoc. 2011, 18, 580–587. [Google Scholar] [CrossRef] [Green Version]
- Cobb, R.; Puri, S.; Wang, D.Z.; Baslanti, T.; Bihorac, A. Knowledge extraction and outcome prediction using medical notes. In Proceedings of the ICML Workshop on Role of Machine Learning in Transforming Healthcare, Atlanta, GA, USA, 20–21 June 2013. [Google Scholar]
- Ni, Y.; Wright, J.; Perentesis, J.; Lingren, T.; Deleger, L.; Kaiser, M.; Kohane, I.; Solti, I. Increasing the efficiency of trial-patient matching: Automated clinical trial eligibility pre-screening for pediatric oncology patients. BMC Med. Inform. Decis. Mak. 2015, 15, 28. [Google Scholar] [CrossRef] [Green Version]
- Garla, V.N.; Brandt, C. Ontology-guided feature engineering for clinical text classification. J. Biomed. Inform. 2012, 45, 992–998. [Google Scholar] [CrossRef]
- Spasić, I.; Livsey, J.; Keane, J.A.; Nenadić, G. Text mining of cancer-related information: Review of current status and future directions. Int. J. Med. Inform. 2014, 83, 605–623. [Google Scholar] [CrossRef]
- Gonzalez-Hernandez, G.; Sarker, A.; O’Connor, K.; Savova, G. Capturing the patient’s perspective: A review of advances in natural language processing of health-related text. Yearb. Med. Inform. 2017, 26, 214. [Google Scholar] [CrossRef]
- Khare, R.; Wei, C.H.; Lu, Z. Automatic extraction of drug indications from FDA drug labels. In Proceedings of the AMIA Annual Symposium Proceedings, Washington, DC, USA, 15–19 November 2014; Volume 2014, p. 787. [Google Scholar]
- Reátegui, R.; Ratté, S. Comparison of MetaMap and cTAKES for entity extraction in clinical notes. BMC Med. Inform. Decis. Mak. 2018, 18, 74. [Google Scholar] [CrossRef] [PubMed]
- Uzuner, Ö. Recognizing obesity and comorbidities in sparse data. J. Am. Med. Inform. Assoc. 2009, 16, 561–570. [Google Scholar] [CrossRef] [Green Version]
- Suominen, H.; Ginter, F.; Pyysalo, S.; Airola, A.; Pahikkala, T.; Salanter, S.; Salakoski, T. Machine learning to automate the assignment of diagnosis codes to free-text radiology reports: A method description. In Proceedings of the ICML/UAI/COLT Workshop on Machine Learning for Health-Care Applications, Helsinki, Filand, 9 July 2008. [Google Scholar]
- Mwangi, B.; Tian, T.S.; Soares, J.C. A review of feature reduction techniques in neuroimaging. Neuroinformatics 2014, 12, 229–244. [Google Scholar] [CrossRef]
- Ngwenya, M. Health Systems Data Interoperability and Implementation. Master’s Thesis, University of South Africa, Pretoria, South Africa, 2018. [Google Scholar]
- Mujtaba, G.; Shuib, L.; Idris, N.; Hoo, W.L.; Raj, R.G.; Khowaja, K.; Shaikh, K.; Nweke, H.F. Clinical text classification research trends: Systematic literature review and open issues. Expert Syst. Appl. 2019, 116, 494–520. [Google Scholar] [CrossRef]
- Sehjal, R.; Harries, V. Awareness of clinical coding: A survey of junior hospital doctors. Br. J. Healthc. Manag. 2016, 22, 310–314. [Google Scholar] [CrossRef]
- Mujtaba, G.; Shuib, L.; Raj, R.G.; Rajandram, R.; Shaikh, K.; Al-Garadi, M.A. Automatic ICD-10 multi-class classification of cause of death from plaintext autopsy reports through expert-driven feature selection. PLoS ONE 2017, 12, e0170242. [Google Scholar]
- Scheurwegs, E.; Cule, B.; Luyckx, K.; Luyten, L.; Daelemans, W. Selecting relevant features from the electronic health record for clinical code prediction. J. Biomed. Inform. 2017, 74, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Scheurwegs, E.; Luyckx, K.; Luyten, L.; Goethals, B.; Daelemans, W. Assigning clinical codes with data-driven concept representation on Dutch clinical free text. J. Biomed. Inform. 2017, 69, 118–127. [Google Scholar] [CrossRef]
- Ferrão, J.C.; Oliveira, M.D.; Janela, F.; Martins, H.M.; Gartner, D. Can structured EHR data support clinical coding? A data mining approach. Health Syst. 2020, 1–24. [Google Scholar] [CrossRef]
- Balakrishnan, S.; Narayanaswamy, R. Feature selection using fcbf in type ii diabetes databases. Int. J. Comput. Internet Manag. 2009, 17, 50–58. [Google Scholar]
- Zhang, W.; Tang, J.; Wang, N. Using the machine learning approach to predict patient survival from high-dimensional survival data. In Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Sehnzhen, China, 15–18 December 2016; pp. 1234–1238. [Google Scholar]
- Buettner, R.; Klenk, F.; Ebert, M. A systematic literature review of machine learning-based disease profiling and personalized treatment. In Proceedings of the 2020 IEEE 44th Annual Computers, Software, and Applications Conference (COMPSAC), Madrid, Spain, 13–17 July 2020. [Google Scholar]
- Yu, L.; Liu, H. Feature selection for high-dimensional data: A fast correlation-based filter solution. In Proceedings of the 20th International Conference on Machine Learning (ICML-03), Washington, DC, USA, 21–24 August 2003; pp. 856–863. [Google Scholar]
- Raza, M.S.; Qamar, U. Understanding and Using Rough Set Based Feature Selection: Concepts, Techniques and Applications; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Goldberg, D.E. Genetic Algorithms; Pearson Education India: Chennai, India, 2006. [Google Scholar]
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Vehtari, A.; Gelman, A.; Gabry, J. Practical Bayesian model evaluation using leave-one-out cross-validation and WAIC. Stat. Comput. 2017, 27, 1413–1432. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, M.; Holländer, N.; Sauerbrei, W. Resampling and cross-validation techniques: A tool to reduce bias caused by model building? Stat. Med. 1997, 16, 2813–2827. [Google Scholar] [CrossRef]
- Saito, T.; Rehmsmeier, M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar] [CrossRef] [Green Version]
- Wolpert, D.H.; Macready, W.G. No free lunch theorems for optimization. IEEE Trans. Evol. Comput. 1997, 1, 67–82. [Google Scholar] [CrossRef] [Green Version]
- Escalante, H.J.; Montes, M.; Sucar, L.E. Particle swarm model selection. J. Mach. Learn. Res. 2009, 10, 405–440. [Google Scholar]
- Kennedy, J.; Eberhart, R. Particle swarm optimization. In Proceedings of the ICNN’95-International Conference on Neural Networks, Perth, Austrilia, 27 November–1 December 1995; Volume 4, pp. 1942–1948. [Google Scholar]
- Pineda, A.L.; Ye, Y.; Visweswaran, S.; Cooper, G.F.; Wagner, M.M.; Tsui, F.R. Comparison of machine learning classifiers for influenza detection from emergency department free-text reports. J. Biomed. Inform. 2015, 58, 60–69. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y. Predicting ICD-9 Codes from Medical Notes–Does the Magic of BERT Applies Here? Available online: https://web.stanford.edu/class/archive/cs/cs224n/cs224n.1204/reports/custom/report25.pdf (accessed on 20 February 2021).
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Zeng, M.; Li, M.; Fei, Z.; Yu, Y.; Pan, Y.; Wang, J. Automatic ICD-9 coding via deep transfer learning. Neurocomputing 2019, 324, 43–50. [Google Scholar] [CrossRef]
- Li, M.; Fei, Z.; Zeng, M.; Wu, F.X.; Li, Y.; Pan, Y.; Wang, J. Automated ICD-9 coding via a deep learning approach. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 16, 1193–1202. [Google Scholar] [CrossRef]
- Malik, S.; Kanwal, N.; Asghar, M.N.; Sadiq, M.A.A.; Karamat, I.; Fleury, M. Data Driven Approach for Eye Disease Classification with Machine Learning. Appl. Sci. 2019, 9, 2789. [Google Scholar] [CrossRef] [Green Version]
- Ananthakrishnan, A.N.; Cai, T.; Savova, G.; Cheng, S.C.; Chen, P.; Perez, R.G.; Gainer, V.S.; Murphy, S.N.; Szolovits, P.; Xia, Z. Improving case definition of Crohn’s disease and ulcerative colitis in electronic medical records using natural language processing: a novel informatics approach. Inflamm. Bowel Dis. 2013, 19, 1411–1420. [Google Scholar] [CrossRef]
- Vukicevic, M.; Radovanovic, S.; Stiglic, G.; Delibasic, B.; Van Poucke, S.; Obradovic, Z. A data and knowledge driven randomization technique for privacy-preserving data enrichment in hospital readmission prediction. In Proceedings of the 5th Workshop on Data Mining for Medicine and Healthcare, Miami, FL, USA, 7 May 2016; Volume 10. [Google Scholar]
- Farkas, R.; Szarvas, G. Automatic construction of rule-based ICD-9-CM coding systems. BMC Bioinform. 2008, 9, S10. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Jamieson, K.; DeSalvo, G.; Rostamizadeh, A.; Talwalkar, A. Hyperband: A novel bandit-based approach to hyperparameter optimization. J. Mach. Learn. Res. 2017, 18, 6765–6816. [Google Scholar]
- Tsoumakas, G.; Katakis, I. Multi-label classification: An overview. Int. J. Data Warehous. Min. (IJDWM) 2007, 3, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Meystre, S.; Haug, P.J. Natural language processing to extract medical problems from electronic clinical documents: Performance evaluation. J. Biomed. Inform. 2006, 39, 589–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miotto, R.; Li, L.; Kidd, B.A.; Dudley, J.T. Deep patient: An unsupervised representation to predict the future of patients from the electronic health records. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sohn, S.; Savova, G.K. Mayo clinic smoking status classification system: Extensions and improvements. In Proceedings of the AMIA Annual Symposium Proceedings, San Francisco, CA, USA, 14–18 November 2009; American Medical Informatics Association: Bethesda, MD, USA, 2009; Volume 2009, p. 619. [Google Scholar]
- Read, J.; Reutemann, P.; Pfahringer, B.; Holmes, G. Meka: A multi-label/multi-target extension to weka. J. Mach. Learn. Res. 2016, 17, 667–671. [Google Scholar]
- Pfaff, E.R.; Crosskey, M.; Morton, K.; Krishnamurthy, A. Clinical Annotation Research Kit (CLARK): Computable Phenotyping Using Machine Learning. JMIR Med. Inform. 2020, 8, e16042. [Google Scholar] [CrossRef]
- Mani, S.; Chen, Y.; Elasy, T.; Clayton, W.; Denny, J. Type 2 diabetes risk forecasting from EMR data using machine learning. In Proceedings of the AMIA Annual Symposium Proceedings, Chicago, IL, USA, 3–7 November 2012; American Medical Informatics Association: Bethesda, MD, USA, 2012; Volume 2012, p. 606. [Google Scholar]
- Skeppstedt, M.; Kvist, M.; Nilsson, G.H.; Dalianis, H. Automatic recognition of disorders, findings, pharmaceuticals and body structures from clinical text: An annotation and machine learning study. J. Biomed. Inform. 2014, 49, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Sohn, S.; Kocher, J.P.A.; Chute, C.G.; Savova, G.K. Drug side effect extraction from clinical narratives of psychiatry and psychology patients. J. Am. Med. Inform. Assoc. 2011, 18, i144–i149. [Google Scholar] [CrossRef] [Green Version]
- Kullo, I.J.; Fan, J.; Pathak, J.; Savova, G.K.; Ali, Z.; Chute, C.G. Leveraging informatics for genetic studies: Use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J. Am. Med. Inform. Assoc. 2010, 17, 568–574. [Google Scholar] [CrossRef] [Green Version]
- Walsh, C.G.; Ribeiro, J.D.; Franklin, J.C. Predicting risk of suicide attempts over time through machine learning. Clin. Psychol. Sci. 2017, 5, 457–469. [Google Scholar] [CrossRef]
- Divita, G.; Luo, G.; Tran, L.; Workman, T.E.; Gundlapalli, A.V.; Samore, M.H. General Symptom Extraction from VA Electronic Medical Notes. Stud. Health Technol. Inform. 2017, 245, 356. [Google Scholar] [PubMed]
- Ghiasvand, O. Disease Name Extraction from Clinical Text Using Conditional Random Fields. Master’s Thesis, University of Wisconsin-Milwaukee, Milwaukee, WI, USA, May 2014. [Google Scholar]
- Guyon, I.; Bennett, K.; Cawley, G.; Escalante, H.J.; Escalera, S.; Ho, T.K.; Macia, N.; Ray, B.; Saeed, M.; Statnikov, A. Design of the 2015 chalearn automl challenge. In Proceedings of the 2015 International Joint Conference on Neural Networks (IJCNN), Killarney, Ireland, 12–16 July 2015; pp. 1–8. [Google Scholar]


| AutoML Platform | Cost | Coding | Location | Dataset | Domain |
|---|---|---|---|---|---|
| Google AutoML | Chargeable | No coding | Cloud | Images Text Tabular | Generic |
| Apple Create ML | Free | Coding needed | Local | Images Text Tabular | Generic |
| Amazon AutoML | Chargeable | No coding | Cloud | Images Text Tabular | Generic |
| Microsoft AutoML | Chargeable | No coding | Cloud | Images Text Tabular | Generic |
| Auto-Sklearn | Free | Coding needed | Local | Tabular | Generic |
| Auto-WEKA | Free | Coding needed | Local | Tabular | Generic |
| Auto-Keras | Free | Coding needed | Local | Tabular | Generic |
| TPOT | Free | Coding needed | Local | Tabular | Generic |
| JADBIO | Chargeable | No coding | Cloud | Tabular | Medical |
| AutoPrognosis | Free | Coding needed | Local | Tabular | Medical |
| Dataset Format | Dataset Type | Disease/ Speciality | Research | AutoML Platform | ||
|---|---|---|---|---|---|---|
| Commercial | Open Source | Health-Related | ||||
| Unstructured | Audio | Hearing Aid | [67] | ✓ | ✕ | ✕ |
| Images | Cancer | [12] | ✓ | ✕ | ✕ | |
| [61] | ✓ | ✕ | ✕ | |||
| Covid-19 | [62] | ✓ | ✕ | ✕ | ||
| Generic | [44] | ✕ | ✕ | ✓ | ||
| [46] | ✕ | ✕ | ✓ | |||
| [63] | ✓ | ✕ | ✕ | |||
| Liver Injury | [64] | ✓ | ✕ | ✕ | ||
| Pachychoroid | [65] | ✓ | ✕ | ✕ | ||
| Structured | Tabular | Alzheimer | [14] | ✕ | ✕ | ✓ |
| BioSignature | [13] | ✕ | ✓ | ✓ | ||
| Brain Age | [58] | ✕ | ✓ | ✕ | ||
| Brain Tumor | [59] | ✕ | ✓ | ✕ | ||
| Cardiac | [25] | ✕ | ✓ | ✓ | ||
| Diabetes | [66] | ✕ | ✓ | ✕ | ||
| Generic | [37] | ✕ | ✓ | ✓ | ||
| [11] | ✕ | ✓ | ✓ | |||
| Metabolic | [60] | ✕ | ✓ | ✕ | ||
| Research | Word Weighting | Word Embedding | Medical Analytics Tools | |||
|---|---|---|---|---|---|---|
| TF-IDF | BOW | Word2vec | GloVe | cTAKES | MetaMap | |
| ML & NLP for clinical notes classification [71] | ✓ | ✓ | ✓ | ✕ | ✓ | ✕ |
| Deep learning evaluation for ICD [20] | ✓ | ✓ | ✓ | ✕ | ✕ | ✕ |
| Clinical text classification [82] | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Labeling clinical text [83] | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Identifying alcohol use [84] | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Automated ICD coding [85] | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Indexing biomedical literature [86] | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Multi-label classification [18] | ✕ | ✓ | ✓ | ✕ | ✕ | ✕ |
| Mental status automated detection [87] | ✕ | ✓ | ✓ | ✕ | ✕ | ✕ |
| ML & NLP for clinical coding [75] | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ |
| ML approach on encoding [89] | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ |
| Feature selection from BOW [88] | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ |
| Medication extraction [74] | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ |
| ICD encoding using deep learning [91] | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ |
| ML models for clinical coding [92] | ✕ | ✕ | ✓ | ✓ | ✕ | ✕ |
| Medical notes classification [93] | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ |
| Embeddings learning from medical notes [94] | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ |
| AI for classifying diagnosis [95] | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ |
| Oncologt patients pre-screening [99] | ✕ | ✕ | ✕ | ✕ | ✓ | ✕ |
| Rules and deep learning comparison [15] | ✕ | ✕ | ✕ | ✕ | ✓ | ✕ |
| Ontology feature engineering [100] | ✕ | ✕ | ✕ | ✕ | ✓ | ✕ |
| Medical notes knowledge extraction [98] | ✕ | ✕ | ✕ | ✕ | ✓ | ✕ |
| Cancer information text mining [101] | ✕ | ✕ | ✕ | ✕ | ✓ | ✕ |
| NLP of health text [102] | ✕ | ✕ | ✕ | ✕ | ✓ | ✕ |
| Drugs indications extraction [103] | ✕ | ✕ | ✕ | ✕ | ✕ | ✓ |
| Radiology reports codes assignment [106] | ✕ | ✕ | ✕ | ✕ | ✕ | ✓ |
| Research | Filter | Wrapper | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IG | MI | SU | GA | FCBF | Forward & Backward Search | LOO | Bootstrap Resampling | Kernel Entropy Inference | ||
| Clinical coding survey [110] | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Autopsy reports classification [111] | ✓ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Clinical coding feature selection [112] | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Ontology feature engineering [100] | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Assigning clinical codes [113] | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Clinical coding with EHR data [114] | ✓ | ✓ | ✕ | ✓ | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ |
| Clinical narrative [21] | ✓ | ✓ | ✕ | ✓ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Health data interoperability [108] | ✕ | ✕ | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ |
| ML diseases profiling [117] | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ |
| Feature selection from BOW [88] | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✓ | ✓ | ✓ |
| Research | Statistical Algorithms | Neural Networks | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RF | LR | DT | SVM | NB | KNN | CNN | RNN | LSTM | GRU | |
| Medical notes classification [93] | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ | ✓ | ✕ | ✓ | ✕ |
| Medication extraction [74] | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | ✓ | ✓ | ✓ | ✕ |
| Automated ICD coding [85] | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ |
| Deep transfer learning for ICD coding [131] | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ |
| ICD coding via deep learning [132] | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ |
| Medical codes explainable prediction [23] | ✕ | ✓ | ✕ | ✓ | ✕ | ✕ | ✓ | ✕ | ✕ | ✓ |
| ML models for clinical coding [92] | ✓ | ✓ | ✓ | ✓ | ✕ | ✕ | ✓ | ✕ | ✓ | ✕ |
| Deep learning evaluation for ICD [20] | ✓ | ✓ | ✕ | ✕ | ✕ | ✕ | ✓ | ✓ | ✓ | ✓ |
| Rules and deep learning comparison [15] | ✓ | ✓ | ✕ | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ |
| AI for classifying diagnosis [95] | ✓ | ✕ | ✕ | ✓ | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ |
| Mental status automated detection [87] | ✓ | ✕ | ✓ | ✓ | ✓ | ✕ | ✓ | ✕ | ✕ | ✕ |
| Automated text classification [17] | ✓ | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ | ✓ | ✕ |
| ML for ICD term encoding [89] | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Eye disease classification with ML [133] | ✓ | ✕ | ✓ | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Autopsy reports classification [111] | ✓ | ✕ | ✓ | ✓ | ✓ | ✓ | ✕ | ✕ | ✕ | ✕ |
| ML classifiers comparison [128] | ✓ | ✓ | ✕ | ✓ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Crohn’s case definition using NLP [134] | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Multi-label classification [18] | ✕ | ✓ | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ |
| Privacy-preserving data enrichment [135] | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | |
| Multi-label classification with DL [16] | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ | ✓ | ✓ | ✓ |
| Rule-based ICD coding [136] | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Diagnosis code assignment [35] | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Ontology feature engineering [100] | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Matching codes to diagnoses [19] | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Medical notes knowledge extraction [98] | ✕ | ✕ | ✕ | ✓ | ✓ | ✓ | ✕ | ✕ | ✕ | ✕ |
| ML for ICD encoding [78] | ✕ | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Determining modification of diagnoses [79] | ✕ | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | ✕ | ✕ | ✕ |
| Research | Recall | Precision | F1 Score | Accuracy | AUC |
|---|---|---|---|---|---|
| Multi-label classification with DL [16] | ✓ | ✓ | ✓ | ✕ | ✕ |
| Automatic recognition of disorders [145] | ✓ | ✓ | ✓ | ✕ | ✕ |
| Diagnosis code assignment [35] | ✓ | ✓ | ✓ | ✕ | ✕ |
| Rules and deep learning comparison [15] | ✓ | ✓ | ✓ | ✕ | ✕ |
| Deep learning evaluation for ICD [20] | ✓ | ✓ | ✓ | ✓ | ✕ |
| Crohn’s case definition using NLP [134] | ✓ | ✓ | ✕ | ✕ | ✕ |
| Drug side effect extraction [146] | ✓ | ✓ | ✓ | ✕ | ✕ |
| Genetic studies informatics leveraging [147] | ✓ | ✓ | ✓ | ✕ | ✕ |
| Smoking status classification [141] | ✓ | ✓ | ✓ | ✕ | ✕ |
| Ontology feature engineering [100] | ✕ | ✕ | ✓ | ✕ | ✕ |
| Multi-label classification [18] | ✕ | ✕ | ✓ | ✕ | ✕ |
| Medication extraction [74] | ✕ | ✕ | ✓ | ✕ | ✕ |
| Matching codes to diagnoses [19] | ✓ | ✓ | ✓ | ✕ | ✕ |
| Automated text classification [17] | ✕ | ✕ | ✓ | ✕ | ✕ |
| Clinical text classification [82] | ✓ | ✓ | ✓ | ✕ | ✕ |
| AI for classifying diagnosis [95] | ✕ | ✕ | ✓ | ✕ | ✓ |
| Automated ICD coding [85] | ✕ | ✕ | ✓ | ✕ | |
| Suicide attempts prediction [148] | ✓ | ✓ | ✕ | ✕ | ✓ |
| Rule-based ICD coding [136] | ✓ | ✓ | ✓ | ✕ | ✕ |
| Radiology reports codes assignment [106] | ✕ | ✕ | ✓ | ✕ | ✕ |
| ICD coding via deep learning [132] | ✓ | ✓ | ✓ | ✕ | ✕ |
| Eye disease classification with ML [133] | ✓ | ✓ | ✓ | ✓ | ✕ |
| Medical codes explainable prediction [23] | ✕ | ✕ | ✓ | ✕ | ✓ |
| ML classifiers comparison [128] | ✓ | ✓ | ✕ | ✓ | ✓ |
| Symptom extraction [149] | ✓ | ✓ | ✓ | ✕ | ✕ |
| Medical problems extraction [139] | ✓ | ✓ | ✓ | ✓ | ✕ |
| ML models for clinical coding [92] | ✓ | ✓ | ✓ | ✕ | ✕ |
| Disease name extraction [150] | ✓ | ✓ | ✓ | ✕ | ✕ |
| Autopsy reports classification [111] | ✓ | ✓ | ✓ | ✓ | ✓ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mustafa, A.; Rahimi Azghadi, M. Automated Machine Learning for Healthcare and Clinical Notes Analysis. Computers 2021, 10, 24. https://0-doi-org.brum.beds.ac.uk/10.3390/computers10020024
Mustafa A, Rahimi Azghadi M. Automated Machine Learning for Healthcare and Clinical Notes Analysis. Computers. 2021; 10(2):24. https://0-doi-org.brum.beds.ac.uk/10.3390/computers10020024
Chicago/Turabian StyleMustafa, Akram, and Mostafa Rahimi Azghadi. 2021. "Automated Machine Learning for Healthcare and Clinical Notes Analysis" Computers 10, no. 2: 24. https://0-doi-org.brum.beds.ac.uk/10.3390/computers10020024






