Uncertainty-Aware Deep Learning-Based Cardiac Arrhythmias Classification Model of Electrocardiogram Signals
Abstract
:1. Introduction
- The ability to outperform the existing results by being able to classify the most extensive set of ECG annotations that are known to us (reaching 20 different imbalanced sets of annotations) in high results.
- The successful incorporation of model uncertainty estimation technique to assist the physician-machine decision making, thus improving the performance regarding the cardiac arrhythmias diagnosis.
2. Preliminaries
2.1. Electrocardiogram (ECG)
2.2. From Recurrent Neural Networks (RNN) to Gated Recurrent Units (GRU)
2.3. Uncertainty in Neural Networks
3. Materials and Methods
3.1. Data Description
3.1.1. MIT-BIH Database
3.1.2. St Petersburg INCART Database
3.1.3. BIDMC Database
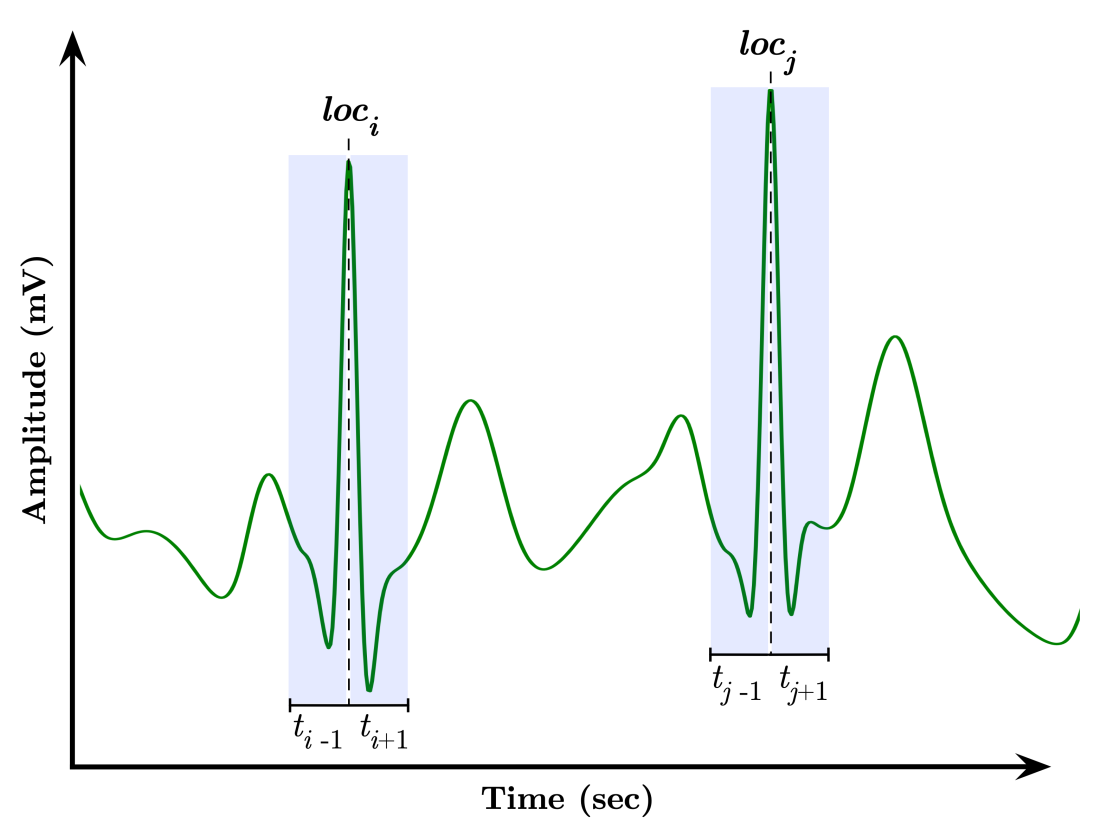
3.2. Signal Segmentation
- Locate QRS locations in the record.
- Extract the corresponding segments using the moving window approach.
- Label the extracted segments using their corresponding annotations.
3.3. Processing Pipeline
3.4. Uncertainty Estimation
4. Experimental Details
4.1. Model Design and Configuration
4.2. Evaluation Metrics
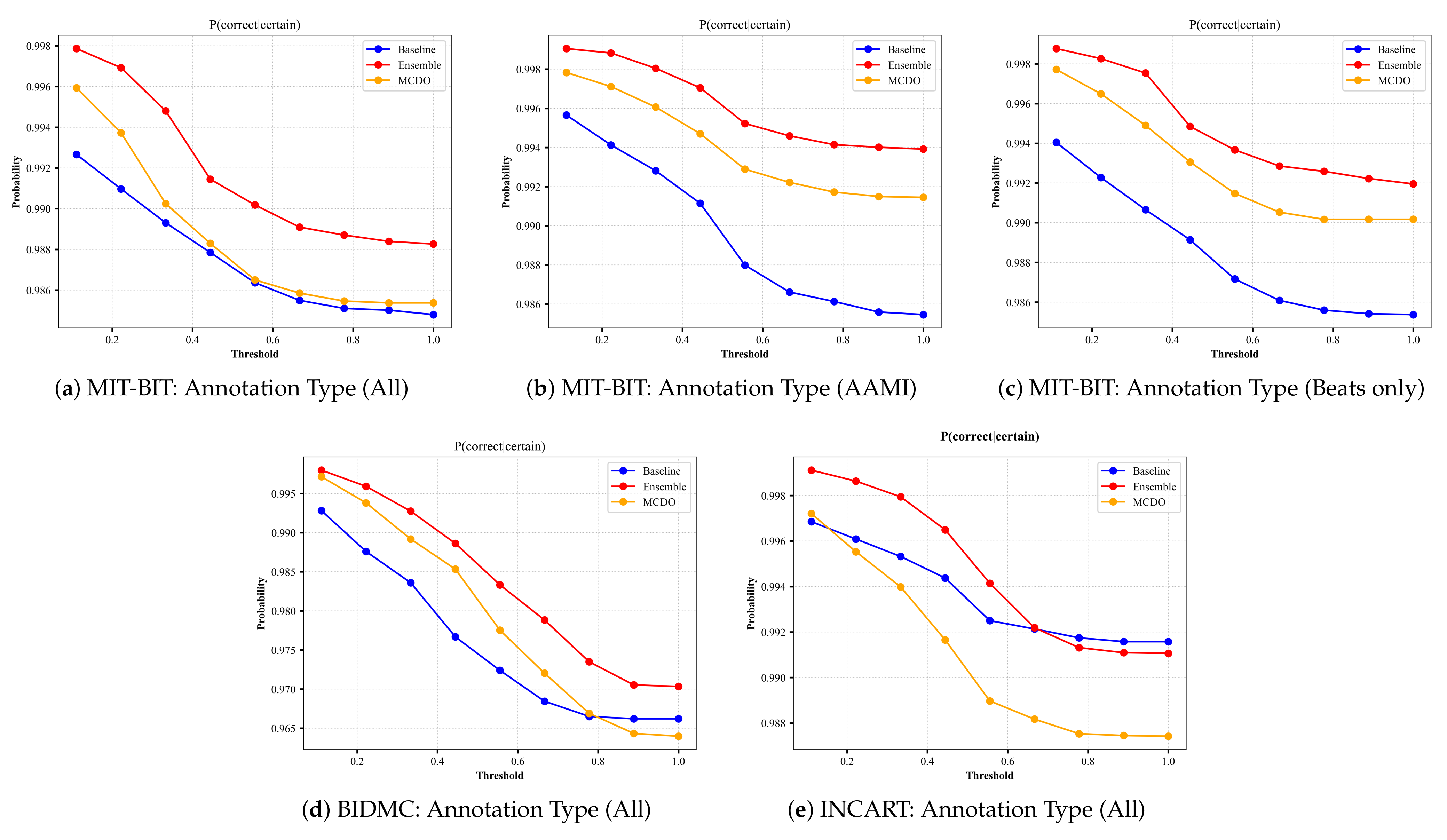
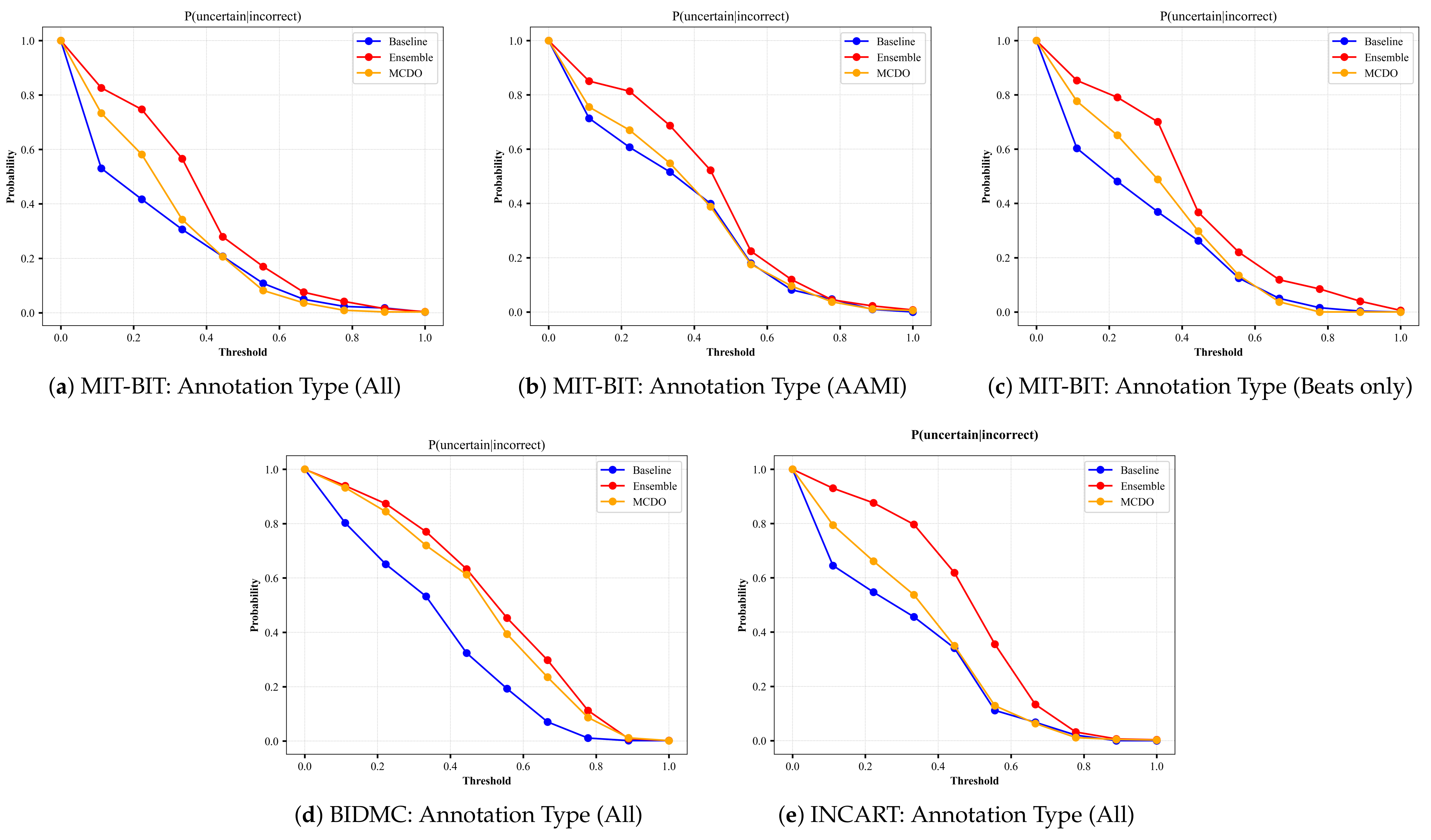
4.3. Probabilistic-Based Metrics
- P(correct|certain) indicates the probability that the model is correct on its outputs given that it is certain about its predictions. This can be calculated as:
- P(uncertain|incorrect) indicates the probability that the model is uncertain about its outputs given that it has produced incorrect predictions.
4.4. Data Oversampling
5. Results and Discussion
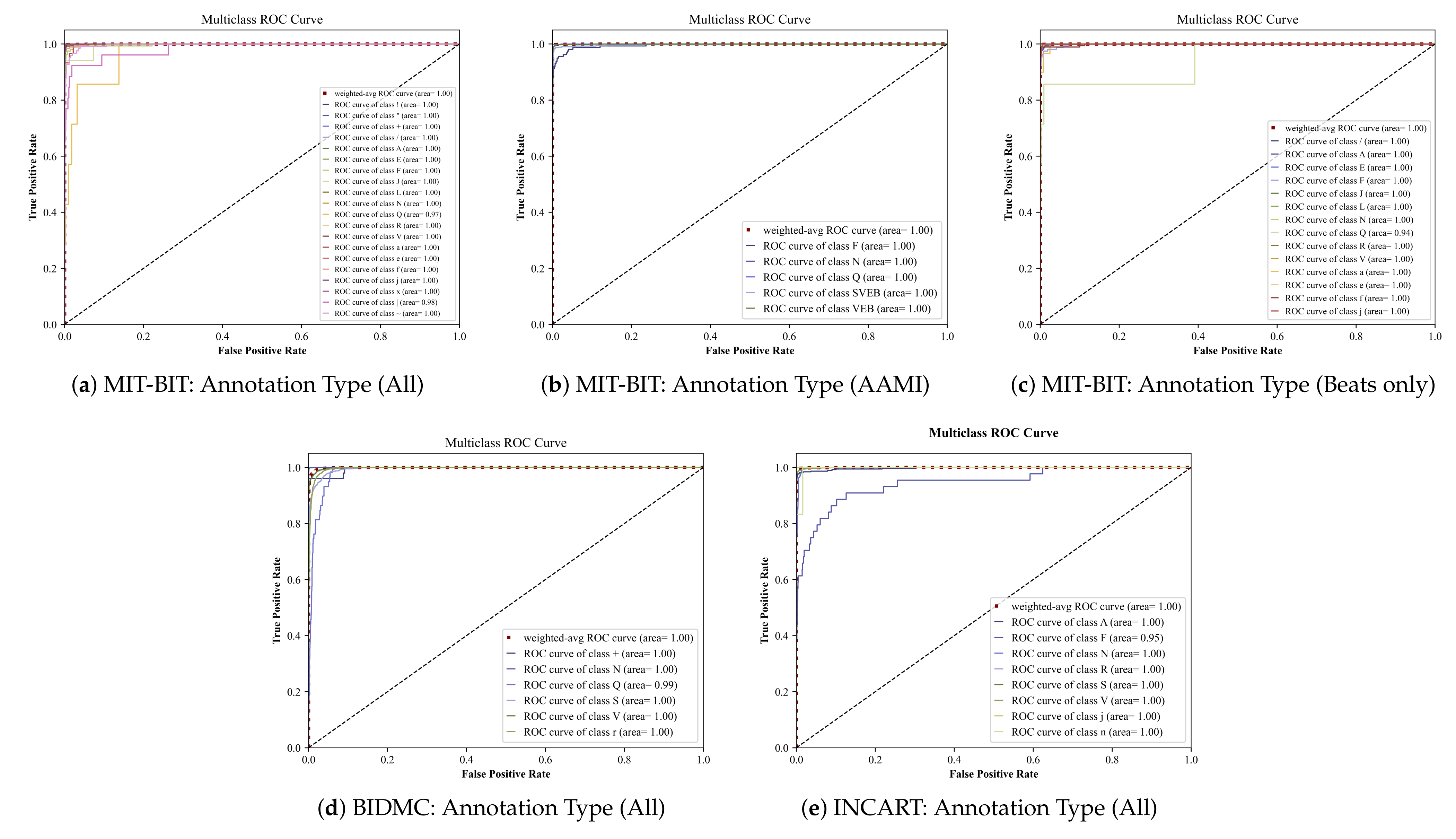
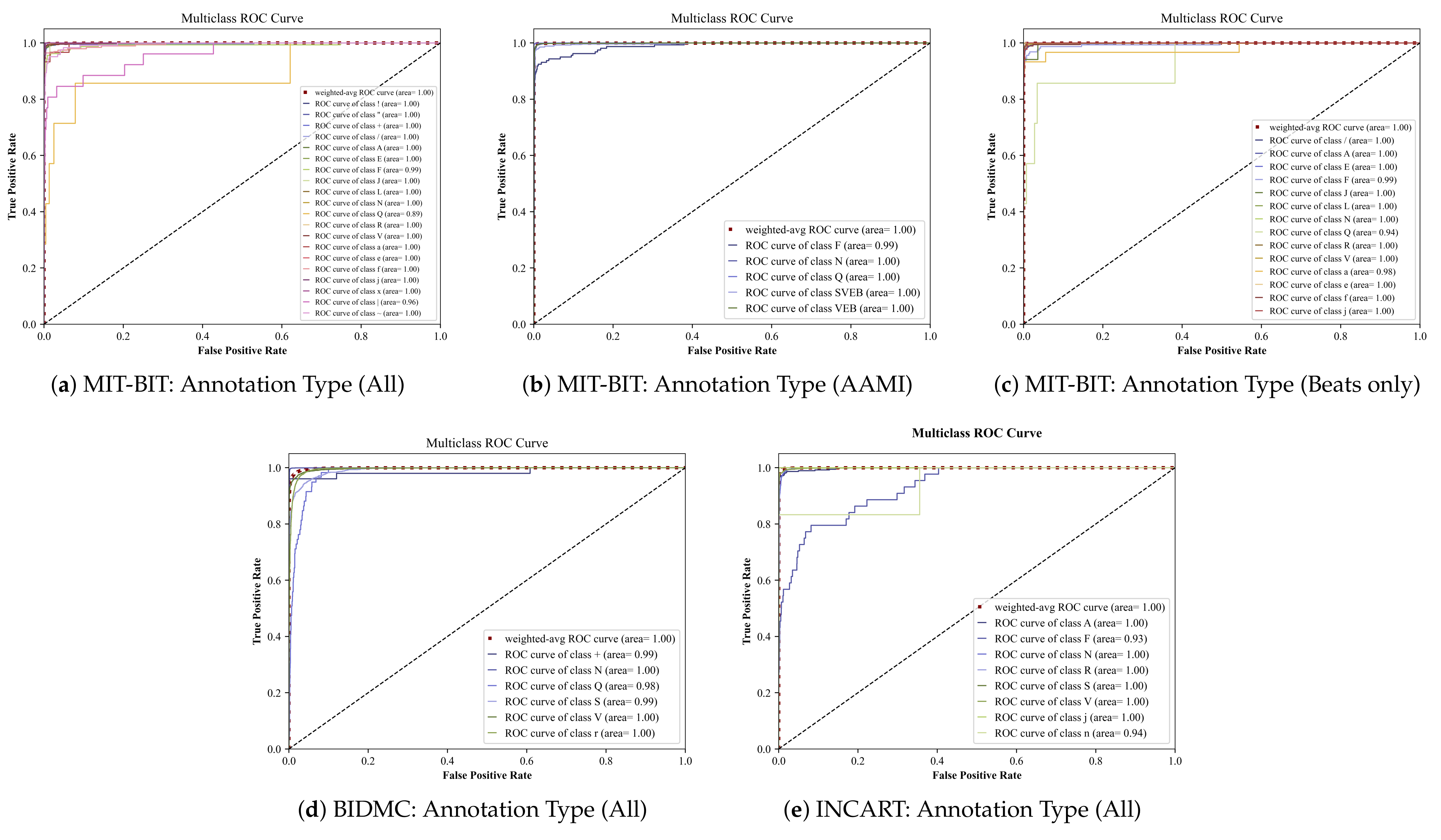
5.1. Deep Ensemble Performance
5.2. Uncertainty Estimation Performance
5.3. Uncertainty Calibration
6. Related Works
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A


References
- Chang, K.C.; Hsieh, P.H.; Wu, M.Y.; Wang, Y.C.; Chen, J.Y.; Tsai, F.J.; Shih, E.S.; Hwang, M.J.; Huang, T.C. Usefulness of machine learning-based detection and classification of cardiac arrhythmias with 12-lead electrocardiograms. Can. J. Cardiol. 2021, 37, 94–104. [Google Scholar] [CrossRef]
- Hijazi, S.; Page, A.; Kantarci, B.; Soyata, T. Machine learning in cardiac health monitoring and decision support. Computer 2016, 49, 38–48. [Google Scholar] [CrossRef]
- Al-Zaiti, S.; Besomi, L.; Bouzid, Z.; Faramand, Z.; Frisch, S.; Martin-Gill, C.; Gregg, R.; Saba, S.; Callaway, C.; Sejdić, E. Machine learning-based prediction of acute coronary syndrome using only the pre-hospital 12-lead electrocardiogram. Nat. Commun. 2020, 11, 1–10. [Google Scholar]
- Giudicessi, J.R.; Schram, M.; Bos, J.M.; Galloway, C.D.; Shreibati, J.B.; Johnson, P.W.; Carter, R.E.; Disrud, L.W.; Kleiman, R.; Attia, Z.I.; et al. Artificial Intelligence–Enabled Assessment of the Heart Rate Corrected QT Interval Using a Mobile Electrocardiogram Device. Circulation 2021, 143, 1274–1286. [Google Scholar] [CrossRef]
- Kashou, A.H.; Noseworthy, P.A. Artificial Intelligence Capable of Detecting Left Ventricular Hypertrophy: Pushing the Limits of the Electrocardiogram? Europace 2020, 22, 338–339. [Google Scholar] [CrossRef]
- Kuncheva, L. Fuzzy Classifier Design; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2000; Volume 49. [Google Scholar]
- Zhang, Y.; Zhou, Z.; Bai, H.; Liu, W.; Wang, L. Seizure classification from EEG signals using an online selective transfer TSK fuzzy classifier with joint distribution adaption and manifold regularization. Front. Neurosci. 2020, 14. [Google Scholar] [CrossRef]
- Postorino, M.N.; Versaci, M. A geometric fuzzy-based approach for airport clustering. Adv. Fuzzy Syst. 2014, 2014. [Google Scholar] [CrossRef] [Green Version]
- Neal, R.M. Bayesian Learning for Neural Networks; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; Volume 118. [Google Scholar]
- Jospin, L.V.; Buntine, W.; Boussaid, F.; Laga, H.; Bennamoun, M. Hands-on Bayesian Neural Networks—A Tutorial for Deep Learning Users. arXiv 2020, arXiv:2007.06823. [Google Scholar]
- Gal, Y.; Ghahramani, Z. Dropout as a bayesian approximation: Representing model uncertainty in deep learning. In Proceedings of the International Conference on Machine Learning, PMLR, New York City, NY, USA, 20–22 June 2016; pp. 1050–1059. [Google Scholar]
- Hein, M.; Andriushchenko, M.; Bitterwolf, J. Why relu networks yield high-confidence predictions far away from the training data and how to mitigate the problem. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 19–20 June 2019; pp. 41–50. [Google Scholar]
- Moon, J.; Kim, J.; Shin, Y.; Hwang, S. Confidence-aware learning for deep neural networks. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual, 13–18 July 2020; pp. 7034–7044. [Google Scholar]
- Gacek, A. An Introduction to ECG Signal Processing and Analysis. In ECG Signal Processing, Classification and Interpretation: A Comprehensive Framework of Computational Intelligence; Gacek, A., Pedrycz, W., Eds.; Springer: London, UK, 2012; pp. 21–46. [Google Scholar] [CrossRef]
- Maglaveras, N.; Stamkopoulos, T.; Diamantaras, K.; Pappas, C.; Strintzis, M. ECG pattern recognition and classification using non-linear transformations and neural networks: A review. Int. J. Med. Inform. 1998, 52, 191–208. [Google Scholar] [CrossRef]
- Rai, H.M.; Trivedi, A.; Shukla, S. ECG signal processing for abnormalities detection using multi-resolution wavelet transform and Artificial Neural Network classifier. Measurement 2013, 46, 3238–3246. [Google Scholar] [CrossRef]
- Morita, H.; Kusano, K.F.; Miura, D.; Nagase, S.; Nakamura, K.; Morita, S.T.; Ohe, T.; Zipes, D.P.; Wu, J. Fragmented QRS as a marker of conduction abnormality and a predictor of prognosis of Brugada syndrome. Circulation 2008, 118, 1697. [Google Scholar] [CrossRef] [Green Version]
- Curtin, A.E.; Burns, K.V.; Bank, A.J.; Netoff, T.I. QRS complex detection and measurement algorithms for multichannel ECGs in cardiac resynchronization therapy patients. IEEE J. Transl. Eng. Health Med. 2018, 6, 1–11. [Google Scholar] [CrossRef]
- Li, C.; Zheng, C.; Tai, C. Detection of ECG characteristic points using wavelet transforms. IEEE Trans. Biomed. Eng. 1995, 42, 21–28. [Google Scholar] [PubMed]
- Chung, J.; Gulcehre, C.; Cho, K.; Bengio, Y. Gated Feedback Recurrent Neural Networks. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 6–11 July 2015; Bach, F., Blei, D., Eds.; PMLR: Lille, France, 2015; Volume 37, pp. 2067–2075. [Google Scholar]
- Xia, Y.; Xiang, M.; Li, Z.; Mandic, D.P. Chapter 12-Echo State Networks for Multidimensional Data: Exploiting Noncircularity and Widely Linear Models. In Adaptive Learning Methods for Nonlinear System Modeling; Comminiello, D., Principe, J.C., Eds.; Butterworth-Heinemann: Oxford, UK, 2018; pp. 267–288. [Google Scholar] [CrossRef]
- Olah, C. Understanding LSTM Networks. Available online: https://colah.github.io/posts/2015-08-Understanding-LSTMs/ (accessed on 17 June 2021).
- Chung, J.; Gulcehre, C.; Cho, K.; Bengio, Y. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv 2014, arXiv:1412.3555. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Gers, F.A.; Schraudolph, N.N.; Schmidhuber, J. Learning precise timing with LSTM recurrent networks. J. Mach. Learn. Res. 2002, 3, 115–143. [Google Scholar]
- Cho, K.; Van Merrinboer, B.; Bahdanau, D.; Bengio, Y. On the properties of neural machine translation: Encoder-decoder approaches. arXiv 2014, arXiv:1409.1259. [Google Scholar]
- Graves, A. Practical variational inference for neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Granada, Spain, 12–15 December 2011; pp. 2348–2356. [Google Scholar]
- Kendall, A.; Gal, Y. What uncertainties do we need in bayesian deep learning for computer vision? arXiv 2017, arXiv:1703.04977. [Google Scholar]
- Welling, M.; Teh, Y.W. Bayesian learning via stochastic gradient Langevin dynamics. In Proceedings of the 28th international conference on machine learning (ICML-11), Citeseer, Bellevue, WA, USA, 28 June–2 July 2011; pp. 681–688. [Google Scholar]
- Hernández-Lobato, J.M.; Adams, R. Probabilistic backpropagation for scalable learning of bayesian neural networks. In Proceedings of the International Conference on Machine Learning, PMLR, Lille, France, 7–9 July 2015; pp. 1861–1869. [Google Scholar]
- Blundell, C.; Cornebise, J.; Kavukcuoglu, K.; Wierstra, D. Weight uncertainty in neural network. In Proceedings of the International Conference on Machine Learning, PMLR, Lille, France, 7–9 July 2015; pp. 1613–1622. [Google Scholar]
- Goldberger, A.; Amaral, L.; Glass, L.; Hausdorff, J.; Ivanov, P.C.; Mark, R.; Mietus, J.; Moody, G.; Peng, C.; Stanley, H. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, E215–E220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef]
- Baim, D.S.; Colucci, W.S.; Monrad, E.S.; Smith, H.S.; Wright, R.F.; Lanoue, A.; Gauthier, D.F.; Ransil, B.J.; Grossman, W.; Braunwald, E. Survival of patients with severe congestive heart failure treated with oral milrinone. J. Am. Coll. Cardiol. 1986, 7, 661–670. [Google Scholar] [CrossRef] [Green Version]
- Research Resource for Complex Physiologic Signals. Available online: https://physionet.org/ (accessed on 17 June 2021).
- Association for the Advancement of Medical Instrumentation. Testing and Reporting Performance Results of Cardiac Rhythm and ST Segment Measurement Algorithms; Association for the Advancement of Medical Instrumentation: Arlington, VA, USA, 1998. [Google Scholar]
- Bleeker, G.B.; Schalij, M.J.; Molhoek, S.G.; Verwey, H.F.; Holman, E.R.; Boersma, E.; Steendijk, P.; Van Der Wall, E.E.; Bax, J.J. Relationship between QRS duration and left ventricular dyssynchrony in patients with end-stage heart failure. J. Cardiovasc. Electrophysiol. 2004, 15, 544–549. [Google Scholar] [CrossRef]
- Sipahi, I.; Carrigan, T.P.; Rowland, D.Y.; Stambler, B.S.; Fang, J.C. Impact of QRS duration on clinical event reduction with cardiac resynchronization therapy: Meta-analysis of randomized controlled trials. Arch. Intern. Med. 2011, 171, 1454–1462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakshminarayanan, B.; Pritzel, A.; Blundell, C. Simple and scalable predictive uncertainty estimation using deep ensembles. arXiv 2016, arXiv:1612.01474. [Google Scholar]
- Leibig, C.; Allken, V.; Ayhan, M.S.; Berens, P.; Wahl, S. Leveraging uncertainty information from deep neural networks for disease detection. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Gal, Y.; Ghahramani, Z. Dropout as a Bayesian approximation: Appendix. arXiv 2015, arXiv:1506.02157. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. 2015. Available online: tensorflow.org (accessed on 17 June 2021).
- Team, K. Simple. Flexible. Powerful. Available online: https://www.myob.com/nz/about/news/2020/simple–flexible–powerful—the-new-myob-essentials (accessed on 17 June 2021).
- Kingma, D.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Aseeri, A. Noise-Resilient Neural Network-Based Adversarial Attack Modeling for XOR Physical Unclonable Functions. J. Cyber Secur. Mobil. 2020, 331–354. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Henne, M.; Schwaiger, A.; Roscher, K.; Weiss, G. Benchmarking Uncertainty Estimation Methods for Deep Learning With Safety-Related Metrics. Available online: http://ceur-ws.org/Vol-2560/paper35.pdf (accessed on 17 June 2021).
- Mukhoti, J.; Gal, Y. Evaluating bayesian deep learning methods for semantic segmentation. arXiv 2018, arXiv:1811.12709. [Google Scholar]
- Lemaître, G.; Nogueira, F.; Aridas, C.K. Imbalanced-learn: A Python Toolbox to Tackle the Curse of Imbalanced Datasets in Machine Learning. J. Mach. Learn. Res. 2017, 18, 1–5. [Google Scholar]
- Ye, C.; Kumar, B.V.; Coimbra, M.T. Combining general multi-class and specific two-class classifiers for improved customized ECG heartbeat classification. In Proceedings of the 21st International Conference on Pattern Recognition (ICPR2012), Tsukuba, Japan, 11–15 November 2012; pp. 2428–2431. [Google Scholar]
- Zhang, Z.; Dong, J.; Luo, X.; Choi, K.S.; Wu, X. Heartbeat classification using disease-specific feature selection. Comput. Biol. Med. 2014, 46, 79–89. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Hannun, A.Y.; Haghpanahi, M.; Bourn, C.; Ng, A.Y. Cardiologist-level arrhythmia detection with convolutional neural networks. arXiv 2017, arXiv:1707.01836. [Google Scholar]
- Acharya, U.R.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M.; Gertych, A.; San Tan, R. A deep convolutional neural network model to classify heartbeats. Comput. Biol. Med. 2017, 89, 389–396. [Google Scholar] [CrossRef]
- He, Z.; Zhang, X.; Cao, Y.; Liu, Z.; Zhang, B.; Wang, X. LiteNet: Lightweight neural network for detecting arrhythmias at resource-constrained mobile devices. Sensors 2018, 18, 1229. [Google Scholar] [CrossRef] [Green Version]
- Jun, T.J.; Nguyen, H.M.; Kang, D.; Kim, D.; Kim, D.; Kim, Y.H. ECG arrhythmia classification using a 2-D convolutional neural network. arXiv 2018, arXiv:1804.06812. [Google Scholar]
- Yang, H.; Wei, Z. Arrhythmia recognition and classification using combined parametric and visual pattern features of ECG morphology. IEEE Access 2020, 8, 47103–47117. [Google Scholar] [CrossRef]
- Carvalho, C.S. A deep-learning classifier for cardiac arrhythmias. arXiv 2020, arXiv:2011.05471. [Google Scholar]




| (a) The number of annotations used by each database (blue colored codes indicate beats-only annotations) | |||||||||||||||||||||||||||
| Database | Records | Annotation Codes Used by the Database | Total | ||||||||||||||||||||||||
| N | A | V | ∼ | | | Q | / | f | + | x | F | j | L | a | J | n | R | [ | ! | ] | E | " | e | r | S | |||
| MIT-BIH | 48 | 74,965 | 2545 | 7126 | 614 | 132 | 33 | 7018 | 982 | 1243 | 193 | 802 | 229 | 8066 | 150 | 83 | 0 | 7251 | 6 | 472 | 6 | 106 | 437 | 16 | 0 | 2 | 112,477 |
| INCART | 75 | 150,253 | 1942 | 19,995 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 219 | 92 | 0 | 0 | 0 | 32 | 3171 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 175,720 |
| BIDMC | 15 | 1,578,105 | 0 | 28,165 | 0 | 0 | 293 | 0 | 0 | 258 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 10,353 | 5314 | 1,622,493 |
| (b) AAMI classes vs. MIT-BIH labels | |||||||||||||||||||||||||||
| AAMI Class | MIT-BIH Beat Annotations | Number of Heartbeats | |||||||||||||||||||||||||
| N | N, L, R, e, j | 90,527 | |||||||||||||||||||||||||
| SVEB | A, a, J, S | 2780 | |||||||||||||||||||||||||
| VEB | V, E | 7232 | |||||||||||||||||||||||||
| Q | P or /, f, Q | 8033 | |||||||||||||||||||||||||
| F | F | 802 | |||||||||||||||||||||||||
| Total | 109,374 | ||||||||||||||||||||||||||
| Dataset | Annotation Type (Size) | Evaluation Metric | |||
|---|---|---|---|---|---|
| Precision | Recall | F1-Score | AUROC | ||
| MIT-BIT | All (20 classes) | 0.9894 | 0.9882 | 0.9887 | 0.9972 |
| Beats only (14 classes) | 0.9921 | 0.9919 | 0.9920 | 0.9955 | |
| AAMI (5 classes) | 0.9939 | 0.9939 | 0.9939 | 0.9987 | |
| BIDMC | All (6 classes) | 0.9756 | 0.9703 | 0.9725 | 0.9960 |
| INCART | All (8 classes) | 0.9940 | 0.9910 | 0.9923 | 0.9922 |
| Dataset | Annotation Type (Size) | Evaluation Metric | |||
|---|---|---|---|---|---|
| Precision | Recall | F1-Score | AUROC | ||
| MIT-BIT | All (20 classes) | 0.9874 | 0.9853 | 0.9862 | 0.9914 |
| Beats only (14 classes) | 0.9908 | 0.9902 | 0.9904 | 0.9932 | |
| AAMI (5 classes) | 0.9917 | 0.9914 | 0.9915 | 0.9970 | |
| BIDMC | All (6 classes) | 0.9718 | 0.9639 | 0.9673 | 0.9925 |
| INCART | All (8 classes) | 0.9921 | 0.9874 | 0.9894 | 0.9834 |
| Authors | Year of Publish | Dataset | Number of Classes | Model Performance |
|---|---|---|---|---|
| Ye et al. [50] | 2012 | MIT-BIH | 5 | Acc: 94% |
| Zhang et al. [51] | 2014 | MIT-BIH | 4 | Acc: 86.66% |
| Rajpurkar et al. [52] | 2017 | Private dataset | 14 | Precision: 80.09% Recall: 82.7% |
| Acharya et al. [53] | 2017 | MIT-BIH | 5 | Noisy set: Acc: 93.47% Sen: 96.01% TNR: 91.64% Noise-free set: Acc: 94.03% Sen: 96.71% TNR: 91.54% |
| He et al. [54] | 2018 | MIT-BIH | 5 | Acc: 98.80% |
| Jun et al. [55] | 2018 | MIT-BIH | 8 | Acc: 99.05% Sen: 99.85% TNR: 99.57% |
| Yang et al. [56] | 2020 | MIT-BIH | 15 | Acc: 97.70% |
| Carvalho [57] | 2020 | MIT-BIH | 13 | Precision: 84.8% Recall: 82.2% |
| This study | 2021 | MIT-BIH | 20 | DE mode: Precision: 98.94% Recall: 98.82% F1: 98.87% MCDO mode: Precision: 98.74% Recall: 98.53% F1: 98.62% |
| BIDMC | 6 | DE mode: Precision: 97.57% Recall: 97.03% F1: 97.25% MCDO mode: Precision: 97.18% Recall: 97.39% F1: 96.73% | ||
| INCART | 8 | DE mode: Precision: 99.4% Recall: 99.1% F1: 99.23% MCDO mode: Precision: 99.21% Recall: 98.74% F1: 96.94% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aseeri, A.O. Uncertainty-Aware Deep Learning-Based Cardiac Arrhythmias Classification Model of Electrocardiogram Signals. Computers 2021, 10, 82. https://0-doi-org.brum.beds.ac.uk/10.3390/computers10060082
Aseeri AO. Uncertainty-Aware Deep Learning-Based Cardiac Arrhythmias Classification Model of Electrocardiogram Signals. Computers. 2021; 10(6):82. https://0-doi-org.brum.beds.ac.uk/10.3390/computers10060082
Chicago/Turabian StyleAseeri, Ahmad O. 2021. "Uncertainty-Aware Deep Learning-Based Cardiac Arrhythmias Classification Model of Electrocardiogram Signals" Computers 10, no. 6: 82. https://0-doi-org.brum.beds.ac.uk/10.3390/computers10060082






