Synthesis and Experimental-Computational Characterization of a Copper/Vanadium Compound with Potential Anticancer Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis and Crystallization
2.2. Theoretical Methodology
3. Results
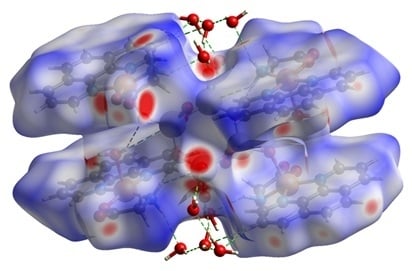
3.1. Structure Description
3.2. Experimental IR and RAMAN Spectra
3.3. 51V Nuclear Magnetic Resonance Spectroscopy
3.4. Visible Spectroscopy
3.5. Cyclic Voltammetry
3.6. Theoretical Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rosenberg, B.; Van Camp, L.; Krigas, T. Inhibition of cell division in Escherichia coli by electrolysis products from a platinum electrode. Nature 1965, 205, 698–699. [Google Scholar] [CrossRef]
- Manzano, C.; Pellei, M.; Tisato, F.; Santini, C. Copper complex as anticancer agents. Anti-Cancer Agents Med. Chem. 2009, 9, 185–211. [Google Scholar] [CrossRef] [PubMed]
- Rabik, C.A.; Dolan, M.E. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat. Rev. 2007, 33, 9–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correia, I.; Roy, S.; Matos, C.P.; Borovic, S.; Butenko, N.; Cavaco, I.; Marques, F.; Lorenzo, J.; Rodríguez, A.; Moreno, V.; et al. Vanadium(IV) and copper(II) complexes of salicylaldimines and aromatic heterocycles: Cytotoxicity, DNA binding, and DNA cleavage properties. J. Inorg. Biochem. 2015, 147, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Denoyer, D.; Clatworthy, S.A.S.; Cater, M.A. Copper Complexes in Cancer Therapy. Met. Ions Life Sci. 2018, 18, 469–506. [Google Scholar]
- Aureliano Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Manzano, C. Advances in Copper Complexes as Anticancer Agents. Chem. Rev. 2014, 815–862. [Google Scholar]
- Li, H.; Wang, J.; Wu, C.; Wang, L.; Chen, Z.S.; Cui, W. The combination of disulfiram and copper for cancer treatment. Drug Discov. Today. 2020. [Google Scholar] [CrossRef]
- Ezhilarasan, D.; Arumugham, M.N. Synthesis, characterization DNA binding and biological activity of Copper(II) complexes with mixed ligands. J. Chem. Biol. Phys. Sci. 2017, 7, 896–905. [Google Scholar]
- Kucková, L.; Jomová, K.; Švorcová, A.; Valko, M.; Segľa, P.; Moncoľ, J.; Kožíšek, J. Synthesis, crystal structure, spectroscopic properties and potential biological activities of salicylate‒neocuproine ternary copper (II) complexes. Molecules 2015, 20, 2115–2137. [Google Scholar] [CrossRef] [Green Version]
- Pages, B.J.; Ang, D.L.; Wright, E.P.; Aldrich-Wright, J.R. Metal complex interactions with DNA. Dalton Trans. 2015, 44, 3505–3526. [Google Scholar] [CrossRef]
- Ng, C.H.; Kong, S.M.; Lian, Y.; Jamil, M.; Sukram, N.; Ahmad, M.; Khoo, A.S.B. Selective anticancer copper(II)-mixed ligand complexes: Targeting of ROS and proteasomes. Metallomics 2014, 6, 892–906. [Google Scholar] [CrossRef]
- Tovar, A.; Ruiz-Ramirez, L.; Campero, A.; Romerosa, A.; Moreno-Esparza, R.; Rosales-Hoz, H. Structural and reactivity studies on 4,4-dimethyl-2,2-bipyridine acetylacetonate copper(II) nitrate (CASIOPEINA III-ia) with methionine, by UV–visible and EPR techniques. J. Inorg. Biochem. 2004, 98, 1045–1053. [Google Scholar] [CrossRef]
- Erxleben, A. Interactions of copper complexes with nucleic acids. Coord. Chem. Rev. 2018, 360, 92–121. [Google Scholar] [CrossRef]
- Baskaran, S.; Krishnan, M.M.; Arumugham, M.N.; Kumar, R.J. DFT analysis and DNA binding, cleavage of copper(II) complexes. Mol. Liq. 2016, 221, 1045–1053. [Google Scholar] [CrossRef]
- Bravo-Gómez, M.E.; García-Ramos, J.C.; Gracia-Mora, I.; Ruiz-Azuara, L. Antiproliferative activity and QSAR study of copper (II) mixed chelate [Cu (N–N)(acetylacetonate)] NO3 and [Cu (N–N)(glycinato)] NO3 complexes,(Casiopeínas®). J. Inorg. Biochem. 2009, 103, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Valdez-Camacho, J.R.; Pérez-Salgado, Y.; Espinoza-Guillén, A.; Gómez-Vidales, V.; Tavira-Montalvan, C.A.; Meneses-Acosta, A.; Leyva, M.A.; Vázquez-Ríos, M.G.; Juaristi, E.; Höpfl, H.; et al. Synthesis, structural characterization and antiproliferative activity on MCF- T 7 and A549 tumor cell lines of [Cu(N-N)(β3-aminoacidate)]NO3 complexes (Casiopeínas®). Inorg. Chim. Acta 2020, 506, 119542. [Google Scholar] [CrossRef]
- Mejia, C.; Ortega-Rosales, S.; Ruiz-Azuara, L. Mechanism of Action of Anticancer Metallodrugs. In Biomedical Applications of Metals; Chapter 10; Springer: Berlin/Heidelberg, Germany, 2018; pp. 213–234. [Google Scholar]
- Treviño, S.; Díaz, A.; Sánchez-Lara, E.; Sanchez-Gaytan, B.L.; Perez-Aguilar, J.M.; González-Vergara, E. Vanadium in Biological Action: Chemical, Pharmacological Aspects, and Metabolic Implications in Diabetes Mellitus. Biol. Trace Elem. Res. 2019, 188, 68–98. [Google Scholar] [CrossRef] [Green Version]
- Bishayee, A.; Waghray, A.; Patel, M.A.; Chatterjee, M. Vanadium in the detection, prevention and treatment of cancer: The in vivo evidence. Cancer Lett. 2010, 294, 1–12. [Google Scholar] [CrossRef]
- Crans, D.; Yang, L.; Haase, A.; Yang, X. Metallo-Drugs: Development and Action of Anticancer Agents. Met. Ions Life Sci. Book 2018, 18, 251–280. [Google Scholar]
- Crans, D.; Smee, J.; Gaidamauskas, E.; Yang, L. The Chemistry and Biochemistry of Vanadium and the Biological Activities Exerted by Vanadium Compounds. Chem. Rev. 2004, 104, 849–902. [Google Scholar] [CrossRef]
- Kioseoglou, E.; Petanidis, S.; Gabriel, C.; Salifoglou, A. The chemistry and biology of vanadium compounds in cancer therapeutics. Coordination. Coord. Chem. Rev. 2015, 301–302, 87–105. [Google Scholar] [CrossRef]
- Novotny, L.; Kombian, S.B. Vanadium: Possible use in cancer chemoprevention and therapy. J. Cancer Res. Updates 2014, 3, 97–102. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, M.; Schwab, M. (Eds.) Encyclopedia of Cancer; Springer: Berlin/Heidelberg, Germany, 2011; pp. 3883–3885. [Google Scholar]
- Shobha, C.; Thulasiram, B.; Aerva, R.; Nagababu, P. Recent Advances in Copper Intercalators as Anticancer Agents. J. Fluoresc. 2018, 28, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Rehder, D. The potentiality of vanadium in medicinal applications. Future Med. Chem. 2012, 4, 1823–1837. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; An, H.; Wang, E.; Sun, C.; Xu, L. Synthesis and structure of a novel one-dimensional vanadate constructed from tetravanadate clusters linked via copper–organic complex moieties: [{Cu(phen)(H2O)}2V4O12]. J. Coord. Chem. 2006, 59, 827–835. [Google Scholar] [CrossRef]
- Kucsera, R.; Joniaková, D.; Zúrková, L. Thermal properties of [MII (phen)3]2V4O12 phen 22H2O (M II = Co, Ni, Cu, phen = 1,10-phenanthroline). J. Therm Anal Calorim. 2004, 78, 263–272. [Google Scholar] [CrossRef]
- Paredes-García, V.; Gaune, S.; Saldías, M.; Garland, M.T.; Baggio, R.; Vega, A.; El Fallah, M.S.; Escuer, A.; Fur, E.L.; Venegas-Yazigi, D.; et al. Solvatomorphs of dimeric transition metal complexes based on the V4O12 cyclic anion as building block: Crystalline packing and magnetic properties. Inorg. Chim. Acta. 2008, 361, 3681–3689. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, X.L.; You, W.S.; Zhao, Y.; Huang, C.Y.; Sun, Z.G. Two novel isomeric complexes supported by vanadates {V4O12}:[{Cu(dpa)2}2V4O12](dpa = 2,2′-dipyridylamine). Inorg. Chem. Commun. 2007, 10, 1465–1468. [Google Scholar] [CrossRef]
- Joniaková, D.; Gyepes, R.; Rakovský, E.; Schwendt, P.; Marek, J.; Mička, Z. Structural variability of copper-1,10-phenanthroline–oxovanadate hybrid inorganic–organic compounds. Polyhedron 2006, 25, 2491–2502. [Google Scholar] [CrossRef]
- Martínez-Valencia, B.; Corona-Motolinia, N.D.; Sánchez-Lara, E.; Noriega, L.; Sánchez-Gaytán, B.L.; Castro, M.E.; Meléndez-Bustamante, F.; González-Vergara, E. Cyclo-tetravanadate bridged copper complexes as potential double bullet pro-metallodrugs for cancer treatment. J. Inorg. Biochem. 2020, 111081. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Mjos, K.D.; Orving, C. Metallodrugs in Medicinal Inorganic Chemistry. Chem. Rev. 2014, 114, 4540–4563. [Google Scholar] [CrossRef] [PubMed]
- Becco, L.; García-Ramos, J.C.; Azuara, L.R.; Gambino, D.; Garat, B. Analysis of the DNA interaction of copper compounds belonging to the Casiopeínas® antitumoral series. Biol. Trace Elem. Res. 2014, 161, 210–215. [Google Scholar] [CrossRef] [PubMed]
- CrysAlis, P.R.O. Agilent Technologies Ltd.: Oxford, UK. 2014. Available online: https://www.agilent.com/cs/library/usermanuals/Public/CrysAlis_Pro_User_Manual.pdf (accessed on 4 June 2020).
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard Puschmann, J.A.K. OLEX2: A complete structure solution, refinement, and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2008, A64, 112–122. [Google Scholar]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Wood, P.A.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Cryst. 2020, 53, 226–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parr, R.G.; Yang, W. Density-Functional Theory of Atoms and Molecules. International Series of Monograph on Chemistry 16; Oxford University Press: New York, NY, USA, 1989; ISBN 978-0-19--987872-7. [Google Scholar]
- Boys, S.F.; Bernardi, F. Calculation of Small Molecular Interactions by Differences of Separate Total Energies—Some Procedures with Reduced Errors. Mol. Phys. 1970, 19, 553. [Google Scholar] [CrossRef]
- Simon, S.; Duran, M.; Dannenberg, J.J. How does basis set superposition error change the potential surfaces for hydrogen bonded dimers? J. Chem. Phys. 1996, 105, 11024–11031. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef]
- Weigend, F. Accurate Coulomb-fitting basis sets for H to Rn. Phys. Chem. Chem. Phys. 2006, 8, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Roy, L.E.; Hay, P.J.; Martin, R.L.J. Revised Basis Sets for the LANL Effective Core Potentials. J. Chem. Theory Comput. 2008, 4, 1029–1031. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, A.; Böhme, M.S.; Dapprich, A.; Gobbi, A.; Höllwarth, V.; Jonas Köhler, K.; Stegmann, R.; Veldkamp, A.; Frenking, G. A set of f-polarization functions for pseudo-potential basis sets of the transition metals SC-Cu, Y-Ag and La-Au. Chem. Phys. Lett. 1993, 208, 111–114. [Google Scholar] [CrossRef]
- Hay, P.J.; Wadt, W.R. Ab initio effective core potentials for molecular calculations. Potentials for the transition metal atoms Sc to Hg. J. Chem. Phys. 1985, 82, 299–310. [Google Scholar] [CrossRef]
- Cossi, M.; Rega, N.; Scalmani, G.; Barone, V. Energies, Structures, and Electronic Properties of Molecules in Solution with the C-PCM Solvation Model. J. Comput. Chem. 2003, 24, 669–681. [Google Scholar] [CrossRef]
- Foster, J.P.; Weinhold, F. Natural hybrid orbitals. J. Am. Chem. Soc. 1980, 102, 7211–7218. [Google Scholar] [CrossRef]
- Reed, A.E.; Curtiss, L.A.; Weinhold, F. Intermolecular interactions from a natural bond orbital, donor-acceptor viewpoint. Chem. Rev. 1988, 88, 899–926. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R. Gaussian 16, Revision, B.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Dennington, R.D., II; Keith, T.A.; Millam, J.M. Gauss View, Version 6.0.16; Semichem, Inc.: Shawnee Mission, UK, 2016. [Google Scholar]
- Turner, M.J.; MacKiinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CRYSTAL EXPLORER; University of Western Australia: Perth, Australia, 2017. Available online: https://crystalexplorer.scb.uwa.edu.au/ (accessed on 4 June 2020).
- Addison, A.W.; Rao, T.N.; Reedijk, J.; Van Rijn, J.; Verschoor, G.C. Synthesis, structure, and spectroscopic properties of copper (II) compounds containing nitrogen–sulphur donor ligands; the crystal and molecular structure of aqua [1,7-bis (N-methylbenzimidazol-2′-yl)-2,6-dithiaheptane] copper (II) perchlorate. J. Chem. Soc. Dalton Trans. 1984, 7, 1349–1356. [Google Scholar] [CrossRef]
- Sharma, R.P.; Ajnesh, S.; Venugopalan, P.; Dansby-Sparks, R.; Xue, Z.L.; Rossetti, S.; Ferretti, V. Stabilization of tetrameric metavanadate ion by tris (1,10-phenanthroline) cobalt(III): Synthesis, spectroscopic, and X-ray structural study of [Co (phen)3]3(V4O12)2Cl 27H2O. J. Coord. Chem. 2010, 63, 3016–3027. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Liang, D.; Wang, M.H. Bis[tris(2,2′-bipyridyl-κ2N,N′)cobalt(II)] cyclo-tetravanadate undecahydrate. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 2013, 69, 138–141. [Google Scholar] [CrossRef]
- Rehder, D. Bioinorganic Vanadium Chemistry; John Wiley & Sons Ltd.: Chichesterm, UK, 2008; Volume 18, p. 59. [Google Scholar]
- Sastry, M.S.; Kesevadas, T.; Rao, G.S.; Sastry, M.D. Phosphate coordination in copper(II) complexes. Proc. Indian Acad. Sci. (Chem. Sci.) 1984, 93, 843–848. [Google Scholar]
- Zhang, W.; Lu, X.; Wang, G.; Cheng, Y.; Zhang, B. Methyl-substituted enhancement of antitumor activity in square-planar metal complex and analysis of DE, DG, CV, UV-vis, and luminescence. New J. Chem. 2015, 39, 4869–4875. [Google Scholar] [CrossRef]
- Sciortino, G.; Maréchal, J.D.; Fábián, I.; Lihi, N.; Garribba, E. Quantitative prediction of electronic absorption spectra of copper (II)–bioligand systems: Validation and applications. J. Inorg. Biochem. 2020, 204, 110953. [Google Scholar] [CrossRef]
- Cakir, S.; Bicer, E. Voltammetric and Spectroscopic Studies of Vanadium (V)-Nicotinamide Interactions at Physiological pH. Turk. J. Chem. 2007, 31, 223–231. [Google Scholar]
- Haque, F.; Rahman, M.S.; Ahmed, E.; Bakshi, P.K.; Shaik, A.A. A Cyclic Voltammetry Study of the Redox Reaction of Cu(II) in the presence of Ascorbic Acid in Different pH Media. Dhaka Univ. J. Sci. 2013, 61, 161–166. [Google Scholar]
- Lu, X.; Zhu, K.; Zhang, M.; Liu, H.; Kang, J. Voltammetric studies of the interaction of transition-metal complexes with DNA. J. Biochem. Biophys. Methods 2002, 52, 189–200. [Google Scholar] [CrossRef]
- Mohamed, M.I.; Shaban, Y.S.; Abd El-Motaled, M.R.; Mohamed, A.A.; Gaber, A.M.M.; Samir, A.E.-S.; Salih, A.-J. Ternary Copper (II) and Nickel (II) chelates of 2,2′-Bipyridyl and glycine: X-ray structures, kinetics, DNA binding, and cleavage activities. J. Mol. Struct. 2019, 1198, 126911. [Google Scholar]
- Janjua, N.K.; Akhter, Z.; Jabeen, F.; Iftikar, B. Cyclic Voltammetric Investigation of Interactions between Bisnitroaromatic Compounds and ds. DNA. J. Korean Chem. Soc. 2014, 58, 153–159. [Google Scholar] [CrossRef] [Green Version]
- Ni, Y.; Lin, D.; Kokot, S. Synchronous fluorescence, UV-visible spectrophotometric, and voltammetric studies of the competitive interaction of bis(1,10-phenanthroline)copper (II) complex and neutral red with DNA. Anal. Biochem. 2006, 352, 231–242. [Google Scholar] [CrossRef]
- Dhakshanamoorthy, S.; Krishnan, M.M.; Arumugham, M.N. Synthesis, Characterisation, DNA Binding/Cleavage, Anticancer and Antimicrobial Activity of Ternary Copper(II) Complexes. Asian J. Res. Chem. 2017, 10, 312–318. [Google Scholar] [CrossRef]
- Zou, X.H.; Ye, B.H.; Li, H.; Zhang, Q.L.; Chao, H.; Liu, J.G.; Ji, L.N.; Li, X.Y. The design of new molecular “light switches” for DNA. J. Biol. Inorg. Chem. 2001, 6, 143–150. [Google Scholar] [CrossRef]
- Chao, H.; Mei, W.J.; Huang, Q.W.; Ji, L.N. NA binding studies of ruthenium(II) complexes containing asymmetric tridentate ligands. J. Inorg. Biochem. 2002, 92, 165–170. [Google Scholar] [CrossRef]
- Kwik, W.L.; Ang, K.P.; Chen, G. Complexes of (2,2′-bipyridyl) copper(II) and (1,10-phenanthroline) copper(II) with some amino acids. J. Inorg. Nucl. Chem. 1980, 42, 303–313. [Google Scholar] [CrossRef]
- Sigman, D.S.; Graham, D.R.; Aurora, V.D.; Stern, A.M. Oxygen-dependent cleavage of DNA by the 1,10-phenanthroline. cuprous complex. Inhibition of Escherichia coli DNA polymerase I. J. Biol. Chem. 1979, 254, 12269–12272. [Google Scholar]
- Zhang, S.; Chun, X.; Chen, Y.; Zhou, J. Synthesis, Crystal Structure and DNA Cleavage Activity of a Ternary Copper(II) Complex of Dipyrido[3,2-d:2′,3′-f]-quinoxaline and Glycine. Chin. J. Chem. 2011, 29, 65–71. [Google Scholar] [CrossRef]
- Alemon-Medina, R.; Brena-Valle, M.; Muñoz-Sánchez, J.L.; Gracia-Mora, M.I.; Ruiz-Azuara, L. Induction of oxidative damage by copper-based antineoplastic drugs (Casiopeínas). Cancer Chemother. Pharmacol. 2007, 60, 219–228. [Google Scholar] [CrossRef]
- Ruiz-Azuara, L. Preparation of new mixed copper aminoacidate complexes from phenylated phenantrolines to be used as “anticancerigenic agents”. U.S. Patent 07 628.628, Re 35,458, 1992. [Google Scholar]
- Rodríguez-Enríquez, S.; Vital-González, P.A.; Flores-Rodríguez, F.L.; Marín-Hernández, A.; Ruiz-Azuara, L.; Moreno-Sánchez, R. Control of cellular proliferation by modulation of oxidative phosphorylation in human and rodent fast-growing tumor cells. Toxicol. Appl. Pharmacol. 2006, 215, 208–217. [Google Scholar] [CrossRef]
- Bravo-Gómez, M.E.; Campero-Peredo, C.; García-Conde, D.; Mosqueira-Santillán, M.J.; Serment-Guerrero, J.; Ruiz-Azuara, L. DNA-Binding Mode of Antitumoral Copper Compounds (Casiopeínas®) and Analysis of its Biological Meaning. Polyhedron 2015, 102, 530–538. [Google Scholar] [CrossRef]
- Serment-Guerrero, J.; Cano-Sanchez, P.; Reyes-Perez, E.; Velazquez-Garcia, F.; Bravo-Gomez, M.E.; Ruiz-Azuara, L. Genotoxicity of the copper antineoplastic coordination complexes Casiopeínas. Toxicol. In Vitro 2011, 25, 1376–1384. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Azuara, L.; Bastian, G.; Bravo-Gómez, M.E.; Cañas, R.C.; Flores-Alamo, M.; Fuentes, I.; Mejia, C.; García-Ramos, J.C.; Serrano, A. Abstract CT408: Phase I study of one mixed chelates copper(II) compound, Casiopeína CasIIIia with antitumor activity and its mechanism of action. Cancer Res. 2014, 74, 19. [Google Scholar]
- Krasnovskaya, O.; Naumov, A.; Guk, D.; Gorelkin, P.; Erofeev, A.; Beloglazkina, E.; Majouga, A. Copper Coordination Compounds as Biologically Active Agents. Int. J. Mol. Sci. 2020, 21, 3965. [Google Scholar] [CrossRef]
- McGivern, T.; Afsharpour, S.; Marmion, C.J. Copper Complexes as Artificial DNA Metallonucleases: From Sigman’s Reagent to Next Generation Anti-Cancer Agent? Inorg. Chim. Acta. 2018, 472, 12–39. [Google Scholar] [CrossRef]
- Griffin, E.; Levina, A.; Lay, P.A. Vanadium(V) tris-3,5-di-tert-butylcatecholato complex: Links between speciation and anti-proliferative activity in human pancreatic cancer cells. J. Inorg. Biochem. 2019, 201, 110815. [Google Scholar] [CrossRef]
- Chaudhary, A.; Singh, A.; Rawat, E.; Singh, R.V. Heterobimetallic Complexes: A Window into Medicinal Chemistry. Chem. Sci. Rev. Lett. 2016, 5, 98–119. [Google Scholar]
- Rozzo, C.; Sanna, D.; Garribba, E.; Serra, M.; Cantara, A.; Palmieri, G.; Pisano, M. Antitumoral effect of vanadium compounds in malignant melanoma cell lines. J. Inorg. Biochem. 2017, 174, 14–24. [Google Scholar] [CrossRef]
- Pisano, M.; Arru, C.; Serra, M.; Galleri, G.; Sanna, D.; Garribba, E.; Palmieri, G.; Rozzo, C. Antiproliferative activity of vanadium compounds: Effects on the major malignant melanoma molecular pathways. Metallomics 2019, 11, 1687–1699. [Google Scholar] [CrossRef]
- Scibior, A.; Pietrzyk, L.; Plewa, Z.; Skiba, A. Vanadium: Risks and possible benefits in the light of a comprehensive overview of its pharmacotoxicological mechanisms and multi-applications with a summary of further research trends. J. Trace Elem. Med. Biol. 2020, 126508. [Google Scholar] [CrossRef]











| Empirical Formula | C56 H72 Cu4 N12 O32 V4 |
|---|---|
| Formula weight | 1883.18 |
| Temperature/K | 293(2) |
| Crystal system | Triclinic |
| Space group | P−1 |
| a/Å b/Å c/Å | 11.9249(4) 13.2626(4) 13.5191(4) |
| α/° β/° γ/° | 109.108(3) 95.685(3) 113.173(3) |
| Volume/Å3 | 1792.27(10) |
| Z | 1 |
| δcalc g/cm3 | 1.745 |
| μ mm−1 | 6.292 |
| F(000) | 956.0 |
| Crystal size/mm3 | 0.303 × 0.14 × 0.052 |
| Radiation | Cu Kα (λ = 1.54184) |
| 2Θ range for data collection/° | 7.172 to 154.71 |
| Index ranges | −15 ≤ h ≤ 14, −16 ≤ k ≤ 16, −16 ≤ l ≤ 17 |
| Reflections collected | 38143 |
| Independent reflections | 7587 [Rint = 0.0568, Rsigma = 0.0496] |
| Data/restraints/parameters | 7587/0/514 |
| Goodness-of-fit on F2 | 1.046 |
| Final R indexes [I > 2σ (I)] | R1 = 0.0380, wR2 = 0.0884 |
| Final R indexes [all data] | R1 = 0.0470, wR2 = 0.0948 |
| Largest diff. peak/hole / e Å−3 | 0.46/−0.36 |
| Atom-Atom | Length/Å | Theoretical/Å |
|---|---|---|
| Cu(1)-O(15) | 1.9441(18) | 1.9343 |
| Cu(1)-N(2) | 2.0127(19) | 2.0304 |
| Cu(1)-O(1) | 2.2371(17) | 2.2260 |
| Cu(1)-N(1) | 2.022(2) | 2.0431 |
| Cu(1)-N(3) | 1.991(2) | 2.0271 |
| Cu(2)-N(5) | 2.0178(19) | 2.0212 |
| Cu(2)-N(4) | 1.9990(18) | 2.0341 |
| Cu(2)-O(7) | 1.931(2) | 1.9483 |
| Cu(2)-N(6) | 1.990(2) | 2.0111 |
| Cu(2)-O(9) | 2.270(2) | 2.3667 |
| V(1)-O(4) | 1.8067(16) | 1.7929 |
| V(1)-O(2) | 1.8002(17) | 1.7922 |
| V(1)-O(1) | 1.6665(15) | 1.6507 |
| V(1)-O(3) | 1.6266(19) | 1.6064 |
| V(2)-O(4) | 1.8166(15) | 1.8268 |
| V(2)-O(2) | 1.8055(17) | 1.8088 |
| V(2)-O(5) | 1.634(2) | 1.6184 |
| V(2)-O(6) | 1.6448(19) | 1.6109 |
| D—H···A | D—H | H···A | D···A | D—H···A |
|---|---|---|---|---|
| O9—H9B···O1 | 0.85 | 1.93 | 2.778 (3) | 177 |
| N3—H3A···O3 | 0.89 | 2.30 | 2.986 (3) | 134 |
| N3—H3A···O13A | 0.89 | 2.31 | 3.050 (6) | 140 |
| N3—H3B···O14 | 0.89 | 2.24 | 3.056 (4) | 153 |
| N3—H3B···O13B | 0.89 | 2.58 | 2.99 (2) | 109 |
| N6—H6A···O14i | 0.89 | 2.44 | 3.281 (4) | 158 |
| N6—H6B···O4 | 0.89 | 2.09 | 2.979 (3) | 172 |
| O9—H9A···O15ii | 0.86 | 2.34 | 3.028 (3) | 137 |
| O9—H9A···O16ii | 0.86 | 2.15 | 2.977 (3) | 161 |
| O9—H9B···O1 | 0.85 | 1.93 | 2.778 (3) | 177 |
| O11—H11A···O12iii | 0.97 | 1.89 | 2.808 (4) | 157 |
| O11—H11B···O16 | 0.94 | 1.90 | 2.813 (3) | 164 |
| O12—H12A···O6ii | 0.85 | 2.04 | 2.876 (3) | 166 |
| O12—H12B···O6iv | 0.85 | 1.99 | 2.833 (3) | 176 |
| O14—H14A···O8Aii | 0.85 | 1.84 | 2.602 (9) | 149 |
| O14—H14A···O8Bii | 0.85 | 1.84 | 2.670 (19) | 167 |
| O14—H14B···O10Av | 0.85 | 1.95 | 2.705 (11) | 147 |
| O14—H14B···O10Bv | 0.85 | 1.92 | 2.663 (15) | 145 |
| O10A—H10B···O5vi | 0.85 | 1.97 | 2.781 (8) | 160 |
| O10B—H10C···O11vii | 0.85 | 2.03 | 2.761 (19) | 144 |
| O10B—H10D···O5vi | 0.85 | 1.92 | 2.755 (16) | 166 |
| O13A—H13A···O3 | 0.85 | 2.50 | 2.982 (4) | 117 |
| O13A—H13A···O5vi | 0.85 | 2.35 | 3.094 (5) | 147 |
| O13A—H13B···O14v | 0.85 | 1.96 | 2.751 (5) | 155 |
| Electronic State | ΔE (kcal mol−1) | Eint (kcal mol−1) |
|---|---|---|
| Singlet | 15.3 | −365.41 |
| Triplet | 0.00 | −410.72 |
| Pentet | 0.10 | −405.20 |
| Interactions | BMetal⋯NoMetal | NPAMetal⋯NoMetal | FVIMetal (au) |
|---|---|---|---|
| Cu(1) ⋯O(15) | 0.341 | 1.073⋯ −0.806 | 0.076 |
| Cu(1) ⋯N(3) | 0.282 | 1.073⋯ −0.974 | 0.076 |
| Cu(1) ⋯N(1) | 0.244 | 1.073⋯ −0.548 | 0.076 |
| Cu(1) ⋯N(2) | 0.243 | 1.073⋯ −0.546 | 0.076 |
| Cu(1) ⋯O(1) | 0.201 | 1.073⋯ −0.635 | 0.076 |
| V(1) ⋯O(1) | 1.661 | 1.884⋯ −0.636 | 2.15 |
| Cu(2) ⋯O(7) | 0.332 | 1.091⋯ −0.800 | 0.085 |
| Cu(2) ⋯N(4) | 0.241 | 1.091⋯ −0.552 | 0.085 |
| Cu(2) ⋯N(5) | 0.238 | 1.091⋯ −0.551 | 0.085 |
| Cu(2) ⋯N(6) | 0.295 | 1.091⋯ −0.945 | 0.085 |
| Cu(2) ⋯O(9) | 0.178 | 1.091⋯ −0.935 | 0.085 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Valencia, B.; Corona-Motolinia, N.D.; Sánchez-Lara, E.; Sánchez-Gaytán, B.L.; Cerro-López, M.; Mendoza, A.; Castro, M.E.; Meléndez-Bustamante, F.J.; González-Vergara, E. Synthesis and Experimental-Computational Characterization of a Copper/Vanadium Compound with Potential Anticancer Activity. Crystals 2020, 10, 492. https://0-doi-org.brum.beds.ac.uk/10.3390/cryst10060492
Martínez-Valencia B, Corona-Motolinia ND, Sánchez-Lara E, Sánchez-Gaytán BL, Cerro-López M, Mendoza A, Castro ME, Meléndez-Bustamante FJ, González-Vergara E. Synthesis and Experimental-Computational Characterization of a Copper/Vanadium Compound with Potential Anticancer Activity. Crystals. 2020; 10(6):492. https://0-doi-org.brum.beds.ac.uk/10.3390/cryst10060492
Chicago/Turabian StyleMartínez-Valencia, Beatriz, Nidia D. Corona-Motolinia, Eduardo Sánchez-Lara, Brenda L. Sánchez-Gaytán, Mónica Cerro-López, Angel Mendoza, María Eugenia Castro, Francisco J. Meléndez-Bustamante, and Enrique González-Vergara. 2020. "Synthesis and Experimental-Computational Characterization of a Copper/Vanadium Compound with Potential Anticancer Activity" Crystals 10, no. 6: 492. https://0-doi-org.brum.beds.ac.uk/10.3390/cryst10060492