Anion Influence on the Packing of 1,3-Bis(4-Ethynyl-3-Iodopyridinium)-Benzene Halogen Bond Receptors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis
2.2. Data Collection and Refinement
2.3. Cambridge Structural Database (CSD) Search
2.4. Computational Analysis
2.5. Reporting Halogen Bond Contacts
3. Results
3.1. Structural Analysis of New Crystals
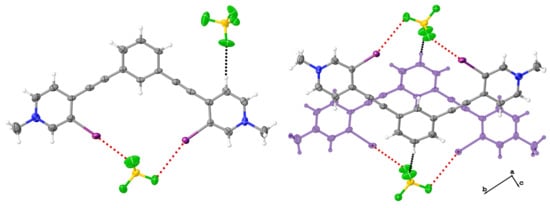
3.1.1. Analysis of 1
3.1.2. Analysis of 2
3.1.3. Analysis of 3•MeOH
3.2. Influence of Anion on Packing Features
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Evans, N.H.; Beer, P.D. Advances in Anion Supramolecular Chemistry: From Recognition to Chemical Applications. Angew. Chem. Int. Ed. 2014, 53, 11716–11754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beer, P.D.; Gale, P.A. Anion Recognition and Sensing: The State of the Art and Future Perspectives. Angew. Chem. Int. Ed. 2001, 40, 486–516. [Google Scholar] [CrossRef]
- Gale, P.A.; Howe, E.N.W.; Wu, X. Anion Receptor Chemistry. Chem. Commun. 2016, 1, 351–422. [Google Scholar] [CrossRef] [Green Version]
- Gale, P.A.; Davis, J.T.; Quesada, R. Anion transport and supramolecular medicinal chemistry. Chem. Soc. Rev. 2017, 46, 2497–2519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Yang, D.; Yang, X.J.; Wu, B. Anion coordination chemistry: From recognition to supramolecular assembly. Coord. Chem. Rev. 2019, 378, 415–444. [Google Scholar] [CrossRef]
- Vilar, R. Anion-Templated Synthesis. Angew. Chem. Int. Ed. 2003, 42, 1460–1477. [Google Scholar] [CrossRef]
- Custelcean, R. Anions in crystal engineering. Chem. Soc. Rev. 2010, 39, 3675. [Google Scholar] [CrossRef]
- Karimi-Jafari, M.; Padrela, L.; Walker, G.M.; Croker, D.M. Creating Cocrystals: A Review of Pharmaceutical Cocrystal Preparation Routes and Applications. Cryst. Growth Des. 2018, 18, 6370–6387. [Google Scholar] [CrossRef]
- Bolla, G.; Nangia, A. Pharmaceutical cocrystals: Walking the talk. Chem. Commun. 2016, 52, 8342–8360. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, Z.; Chen, Z.; Zhang, Q. Organic Cocrystals: Beyond Electrical Conductivities and Field-Effect Transistors (FETs). Angew. Chem. Int. Ed. 2019, 2–18. [Google Scholar]
- Zhang, C.; Xiong, Y.; Jiao, F.; Wang, M.; Li, H. Redefining the Term of “Cocrystal” and Broadening Its Intention. Cryst. Growth Des. 2019, 19, 1471–1478. [Google Scholar] [CrossRef]
- Gryl, M.; Kozieł, M.; Stadnicka, K.M. A proposal for coherent nomenclature of multicomponent crystals. Acta Crystallogr. Sect. B 2019, 75, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Grothe, E.; Meekes, H.; Vlieg, E.; ter Horst, J.H.; de Gelder, R. Solvates, Salts, and Cocrystals: A Proposal for a Feasible Classification System. Cryst. Growth Des. 2016, 16, 3237–3243. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Cabeza, A.J. Acid–base crystalline complexes and the pKa rule. Cryst. Eng. Comm. 2012, 14, 6362. [Google Scholar] [CrossRef]
- Lieffrig, J.; Jeannin, O.; Frąckowiak, A.; Olejniczak, I.; Świetlik, R.; Dahaoui, S.; Aubert, E.; Espinosa, E.; Auban-Senzier, P.; Fourmigué, M. Charge-Assisted Halogen Bonding: Donor-Acceptor Complexes with Variable Ionicity. Chem. Eur. J. 2013, 19, 14804–14813. [Google Scholar] [CrossRef] [PubMed]
- Walter, S.M.; Kniep, F.; Rout, L.; Schmidtchen, F.P.; Herdtweck, E.; Huber, S.M. Isothermal Calorimetric Titrations on Charge-Assisted Halogen Bonds: Role of Entropy, Counterions, Solvent, and Temperature. J. Am. Chem. Soc. 2012, 134, 8507–8512. [Google Scholar] [CrossRef] [PubMed]
- Desiraju, G.R.; Ho, P.S.; Kloo, L.; Legon, A.C.; Marquardt, R.; Metrangolo, P.; Politzer, P.; Resnati, G.; Rissanen, K. Definition of the halogen bond (IUPAC Recommendations 2013). Pure Appl. Chem. 2013, 85, 1711–1713. [Google Scholar] [CrossRef]
- Brown, A.; Beer, P.D. Halogen bonding anion recognition. Chem. Commun. 2016, 52, 8645–8658. [Google Scholar] [CrossRef] [PubMed]
- Cavallo, G.; Metrangolo, P.; Milani, R.; Pilati, T.; Priimagi, A.; Resnati, G.; Terraneo, G. The Halogen Bond. Chem. Rev. 2016, 116, 2478–2601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tepper, R.; Schubert, U.S. Halogen Bonding in Solution: Anion Recognition, Templated Self-Assembly, and Organocatalysis. Angew. Chem. Int. Ed. 2018, 57, 6004–6016. [Google Scholar] [CrossRef]
- Rowe, R.K.; Ho, P.S. Relationships between hydrogen bonds and halogen bonds in biological systems. Acta Crystallogr. Sect. B 2017, 73, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Auffinger, P.; Hays, F.A.; Westhof, E.; Ho, P.S. Halogen bonds in biological molecules. Proc. Natl. Acad. Sci. USA 2004, 101, 16789–16794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kniep, F.; Jungbauer, S.H.; Zhang, Q.; Walter, S.M.; Schindler, S.; Schnapperelle, I.; Herdtweck, E.; Huber, S.M. Organocatalysis by Neutral Multidentate Halogen-Bond Donors. Angew. Chem. Int. Ed. 2013, 52, 7028–7032. [Google Scholar] [CrossRef] [PubMed]
- Walter, S.M.; Kniep, F.; Herdtweck, E.; Huber, S.M. Halogen-Bond-Induced Activation of a Carbon-Heteroatom Bond. Angew. Chem. Int. Ed. 2011, 50, 7187–7191. [Google Scholar] [CrossRef] [PubMed]
- Jungbauer, S.H.; Huber, S.M. Cationic Multidentate Halogen-Bond Donors in Halide Abstraction Organocatalysis: Catalyst Optimization by Preorganization. J. Am. Chem. Soc. 2015, 137, 12110–12120. [Google Scholar] [CrossRef] [PubMed]
- Dumele, O.; Schreib, B.; Warzok, U.; Trapp, N.; Schalley, C.A.; Diederich, F. Halogen-Bonded Supramolecular Capsules in the Solid State, in Solution, and in the Gas Phase. Angew. Chem. Int. Ed. 2017, 56, 1152–1157. [Google Scholar] [CrossRef] [PubMed]
- Dumele, O.; Trapp, N.; Diederich, F. Halogen Bonding Molecular Capsules. Angew. Chem. Int. Ed. 2015, 54, 12339–12344. [Google Scholar] [CrossRef] [PubMed]
- Beyeh, N.K.; Pan, F.; Rissanen, K. A Halogen-Bonded Dimeric Resorcinarene Capsule. Angew. Chem. 2015, 127, 7411–7415. [Google Scholar] [CrossRef]
- Massena, C.J.; Wageling, N.B.; Decato, D.A.; Martin Rodriguez, E.; Rose, A.M.; Berryman, O.B. A Halogen-Bond-Induced Triple Helicate Encapsulates Iodide. Angew. Chem. Int. Ed. 2016, 55, 12398–12402. [Google Scholar] [CrossRef]
- Massena, C.J.; Decato, D.A.; Berryman, O.B. A long-lived Halogen-Bonding anion triple helicate accommodates rapid guest exchange. Angew. Chem. Int. Ed. 2018, 57, 16109–16113. [Google Scholar] [CrossRef] [PubMed]
- Metrangolo, P.; Pilati, T.; Terraneo, G.; Biella, S.; Resnati, G. Anion coordination and anion-templated assembly under halogen bonding control. Cryst. Eng. Comm. 2009, 11, 1187. [Google Scholar] [CrossRef]
- Cavallo, G.; Metrangolo, P.; Pilati, T.; Resnati, G.; Sansotera, M.; Terraneo, G. Halogen bonding: A general route in anion recognition and coordination. Chem. Soc. Rev. 2010, 39, 3772. [Google Scholar] [CrossRef] [PubMed]
- Carroll, C.N.; Naleway, J.J.; Haley, M.M.; Johnson, D.W. Arylethynyl receptors for neutral molecules and anions: Emerging applications in cellular imaging. Chem. Soc. Rev. 2010, 39, 3875. [Google Scholar] [CrossRef] [PubMed]
- Vonnegut, C.L.; Tresca, B.W.; Johnson, D.W.; Haley, M.M. Ion and Molecular Recognition Using Aryl-Ethynyl Scaffolding. Chem. Asian J. 2015, 10, 522–535. [Google Scholar] [CrossRef] [PubMed]
- Leininger, S.; Olenyuk, B.; Stang, P.J. Self-Assembly of discrete cyclic nanostructures mediated by transition metals. Chem. Rev. 2000, 100, 853–908. [Google Scholar] [CrossRef]
- Massena, C.J.; Riel, A.M.S.; Neuhaus, G.F.; Decato, D.A.; Berryman, O.B. Solution and solid-phase halogen and C-H hydrogen bonding to perrhenate. Chem. Commun. 2015, 51, 1417–1420. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Riel, A.M.S.; Berryman, O.B. Solvatochromism and fluorescence response of a halogen bonding anion receptor. New J. Chem. 2018, 42, 10489–10492. [Google Scholar] [CrossRef]
- Riel, A.M.S.; Jessop, M.J.; Decato, D.A.; Massena, C.J.; Nascimento, V.R.; Berryman, O.B. Experimental investigation of halogen-bond hard-soft acid-base complementarity. Acta Crystallogr. Sect. B 2017, 73, 203–209. [Google Scholar] [CrossRef]
- Riel, A.M.S.; Decato, D.A.; Sun, J.; Massena, C.J.; Jessop, M.J.; Berryman, O.B. The intramolecular hydrogen bonded-halogen bond: A new strategy for preorganization and enhanced binding. Chem. Sci. 2018, 9, 5828–5836. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SADABS: Area Detector Absorption Correction; University of Gottingen: Alemanha, Germany, 1996. [Google Scholar]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Crystallogr. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Bruker, APEX2; Bruker AXS Inc: Madison, WI, USA, 2007.
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Spek, A.L. PLATON SQUEEZE: A tool for the calculation of the disordered solvent contribution to the calculated structure factors. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Bruno, I.J.; Cole, J.C.; Edgington, P.R.; Kessler, M.; Macrae, C.F.; Mccabe, P.; Pearson, J.; Taylor, R. Structural Science New software for searching the Cambridge Structural Database and visualizing crystal structures. Acta Cryst. 2002, 58, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge structural database. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 2016, B72, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Wolff, S.K.; Grimwood, D.J.; McKinnon, J.J.; Turner, M.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer (Version 3.1); University of Western Australia: Crawley, Austrália, 2012. [Google Scholar]
- Spackman, M.A.; Byrom, P.G. A novel definition of a molecule in a crystal. Chem. Phys. Lett. 1997, 267, 215–220. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Alvarez, S. A cartography of the van der Waals territories. Dalt. Trans. 2013, 42, 8617. [Google Scholar] [CrossRef]
- Sirohiwal, A.; Hathwar, V.R.; Dey, D.; Regunathan, R.; Chopra, D. Characterization of fluorine-centred ‘F...O’ σ-hole interactions in the solid state. Acta Crystallogr. Sect. B 2017, 73, 140–152. [Google Scholar] [CrossRef] [PubMed]
- Dikundwar, A.G.; Row, T.N.G. Evidence for the “Amphoteric” Nature of Fluorine in Halogen Bonds: An Instance of Cl···F Contact. Cryst. Growth Des. 2012, 12, 1713–1716. [Google Scholar] [CrossRef]
- Metrangolo, P.; Murray, J.S.; Pilati, T.; Politzer, P.; Resnati, G.; Terraneo, G. The fluorine atom as a halogen bond donor, viz. a positive site. Cryst. Eng. Comm. 2011, 13, 6593. [Google Scholar] [CrossRef]
- Alkorta, I.; Elguero, J.; Del Bene, J.E. Boron as an Electron-Pair Donor for B…Cl Halogen Bonds. Chem. Phys. Chem. 2016, 17, 3112–3119. [Google Scholar] [CrossRef]
- Zhuo, H.; Yu, H.; Li, Q.; Li, W.; Cheng, J. Some measures for mediating the strengths of halogen bonds with the B-B bond in diborane(4) as an unconventional halogen acceptor. Int. J. Quantum Chem. 2014, 114, 128–137. [Google Scholar] [CrossRef]
- Kitaigoradskii, A.I. Organic Chemical Crystallography; Consultants Bureau: New York, NY, USA, 1961. [Google Scholar]
- Corpinot, M.K.; Bučar, D.K. A Practical Guide to the Design of Molecular Crystals. Cryst. Growth Des. 2019, 19, 1426–1453. [Google Scholar] [CrossRef]
- Riel, A.M.S.; Decato, D.A.; Berryman, O.B. Protonation and Alkylation Induced Multidentate C–H···Anion Binding to Perrhenate. Cryst. Growth Des. 2016, 16, 974–980. [Google Scholar] [CrossRef]
- Berryman, O.B.; Johnson, C.A.; Vonnegut, C.L.; Fajardo, K.A.; Zakharov, L.N.; Johnson, D.W.; Haley, M.M. Solid-State Examination of Conformationally Diverse Sulfonamide Receptors Based on Bis(2-anilinoethynyl)pyridine, -Bipyridine, and -Thiophene. Cryst. Growth Des. 2015, 15, 1502–1511. [Google Scholar] [CrossRef] [Green Version]
- Lohrman, J.A.; Deng, C.-L.; Shear, T.A.; Zakharov, L.N.; Haley, M.M.; Johnson, D.W. Methanesulfonyl-polarized halogen bonding enables strong halide recognition in an arylethynyl anion receptor. Chem. Commun. 2019, 55, 1919–1922. [Google Scholar] [CrossRef]
- Molčanov, K.; Milašinović, V.; Kojić-Prodić, B. Contribution of Different Crystal Packing Forces in π-Stacking: From Noncovalent to Covalent Multicentric Bonding. Cryst. Growth Des. 2019, 19, 5967–5980. [Google Scholar] [CrossRef]










| Crystal Parameter | 1 | 2 | 3•MeOH |
|---|---|---|---|
| CCDC | 1,936,528 | 1,936,530 | 1,936,529 |
| empirical formula | C22H16B2F8I2N2 | C22H16I2N2O8S2 | C23H20I2N4O7 |
| formula weight | 735.79 | 754.29 | 718.23 |
| temp (K) | 100 | 100 | 150 |
| crystal system | monoclinic | monoclinic | triclinic |
| space group | P21/c | I2/a | P |
| a (Å) | 7.0124(7) | 14.4133(6) | 7.2630(7) |
| b (Å) | 32.096(3) | 12.5563(5) | 11.0928(11) |
| c (Å) | 11.4846(12) | 33.0305(16) | 16.7314(17) |
| α (°) | 90 | 90 | 81.574(3) |
| β (°) | 103.063(2) | 91.905(2) | 77.700(3) |
| γ (°) | 90 | 90 | 78.539(3) |
| V (Å3) | 2517.9(4) | 5974.5(4) | 1283.1(2) |
| Z | 4 | 8 | 2 |
| Dc (g/cm3) | 1.941 | 1.677 | 1.859 |
| μ (mm−1) | 2.572 | 2.288 | 2.501 |
| F (000) | 1400.0 | 2912.0 | 696.0 |
| crystal size (mm) | 0.30 × 0.04 × 0.03 | 0.35 × 0.03 × 0.02 | 0.35 × 0.05 × 0.03 |
| 2Θ max (°) | 50.05 | 52.744 | 54.968 |
| no. of reflections | 33,888 | 106,893 | 42,751 |
| no. of independent reflections | 4444 | 6106 | 5904 |
| Rint | 0.0427 | 0.0454 | 0.0368 |
| GOF | 1.460 | 1.062 | 1.055 |
| R1 (I > 2σ(I)) | 0.0634 | 0.0404 | 0.0335 |
| wR2 (I > 2σ(I)) | 0.1482 | 0.0960 | 0.0801 |
| max/min residual e− density (e Å−3) | 1.62/−1.20 | 2.81/−1.21 | 1.47/−0.55 |
| Compound | Interaction | C-I···LB Distance (Å) | Reduction Ratio* | C-I···LB Angle (°) |
|---|---|---|---|---|
| 1 | C-I···F (I1-F4) | 3.396(7) | 0.97 | 161.2(3) |
| C-I···F (I2-F1) | 3.100(8) | 0.89 | 173.5(3) | |
| C-I···B (I2-B1) | 3.767(13) | 0.95 | 164.1(3) | |
| 3•MeOH | C-I···O (I2-O1) | 2.903(3) | 0.82 | 176.16(13) |
| C-I···O (I1-O1) | 2.965(3) | 0.84 | 175.19(14) | |
| 2 | C-I···O (I1-O1) | 3.005(5) | 0.85 | 167.99(14) |
| C-I···O (I2-O8) | 3.244(13) | 0.92 | 169.1(3) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Decato, D.A.; Riel, A.M.S.; Berryman, O.B. Anion Influence on the Packing of 1,3-Bis(4-Ethynyl-3-Iodopyridinium)-Benzene Halogen Bond Receptors. Crystals 2019, 9, 522. https://0-doi-org.brum.beds.ac.uk/10.3390/cryst9100522
Decato DA, Riel AMS, Berryman OB. Anion Influence on the Packing of 1,3-Bis(4-Ethynyl-3-Iodopyridinium)-Benzene Halogen Bond Receptors. Crystals. 2019; 9(10):522. https://0-doi-org.brum.beds.ac.uk/10.3390/cryst9100522
Chicago/Turabian StyleDecato, Daniel A., Asia Marie S. Riel, and Orion B. Berryman. 2019. "Anion Influence on the Packing of 1,3-Bis(4-Ethynyl-3-Iodopyridinium)-Benzene Halogen Bond Receptors" Crystals 9, no. 10: 522. https://0-doi-org.brum.beds.ac.uk/10.3390/cryst9100522