1. Introduction
X-ray structure analysis has an irreplaceable role in the determination of crystal structures of organic compounds, as supported by almost a million entries in the Cambridge Structure Database (CSD) [
1]. Regarding single crystal X-ray analysis, the final structure is obtained in several steps performed in a specific order [
2]. The chosen single crystal is mounted in a diffractometer, and within a few minutes, several diffraction frames are acquired. Based on the reflection positions, unit cell parameters are calculated, revealing the crystal system. The accuracy of the cell parameters rises with the number of acquired reflections. Subsequently, a particular space group of the system is proposed. With a sufficient number of reflections, the unit cell parameters and the appropriate space group, the crystal structure can be solved. Nowadays, there are plenty of methods enabling structure solution. The original heavy atom methods [
3] have been progressively substituted by direct methods [
4]. The X-ray beam interacts with the electrons and diffracts on periodic electron density in a given direction. The initial structure solution always consists of a coarse skeleton of heavy atoms, as lighter atoms scatter weakly due to the low number of electrons. Light atoms have to be localized and refined at the very end of structure refinement. It has to be pointed out that even the positions of the lightest atoms, the hydrogen atoms, can be localized from residual electron density maps using good-quality diffraction data. The position of the hydrogen atom indicates the location of its electron, which is pulled towards the bonding partner; therefore, it does not coincide with the position of the hydrogen nucleus. Having just one electron, the contribution of hydrogen atoms to the diffraction pattern is rather small.
An alternative experimental technique for the characterization of solids is solid-state NMR spectroscopy (SS-NMR). NMR-based crystallography is a rapidly developing field generating around one thousand research articles per year, according to Web of Science (WOS) [
5]. In contrast to X-ray diffraction, hydrogen atoms are the most sensitive for NMR spectroscopy. Furthermore, SS-NMR does not require a long-range order in the studied materials, and is therefore suitable for characterization of disordered and amorphous samples [
6,
7,
8,
9,
10]. X-ray diffraction and SS-NMR are thus complementary techniques in many respects. The progress of NMR crystallography in recent years can mostly be attributed to the development of fast and reliable computational techniques (mostly based on density functional theory) that enable the linking of experimentally observable parameters with structural models [
11,
12,
13].
However, the main challenge of NMR crystallography is represented by direct structure elucidation. Although it was demonstrated that
de novo crystal structure predictions are possible for NMR crystallography, all current procedures place high demands on computer time, because the NMR parameters of a large set of initially predicted crystal structures have to be calculated and compared with the experimental data [
14,
15]. Therefore, the majority of current NMR crystallography procedures use crystallographic parameters as an input, namely, cell lattice parameters and a space group, which are generally inaccessible by NMR methods.
In this paper, the hypothetical possibility of reconstructing the molecular packing network and determining the crystallographic parameters using just NMR data is explored. Structural data of
l-histidine·HCl·H
2O are used for a case simulation, considering that cell parameters could not be determined from X-ray powder diffraction for some reason, which is quite common for monoclinic and triclinic systems. This particular system was chosen because the recent pioneering work of Nishiyama and coworkers has demonstrated that intermolecular distances can be obtained with SS-NMR experiments [
16,
17]. The newly developed approaches were demonstrated on a crystalline sample of
l-histidine·HCl·H
2O. In addition to 10 intramolecular hydrogen interactions, the
1H DQ-SQ correlation spectrum collected in ref. [
16] also revealed 7 intermolecular hydrogen-hydrogen interactions. Moreover, two intermolecular contacts between nitrogen and hydrogen atoms were observed in the 2D
14N(SQ)-
1H(SQ) spectrum. Altogether, 9 intermolecular distances were described in ref. [
16].
Considering that quantification of intermolecular distances will be feasible experimentally in the future, the procedure for the derivation of standard crystallographic parameters from such data was attempted here. Solid state NMR can easily determine the number of non-equivalent molecules in the asymmetric unit (
Z’) [
18]. In accordance with the experiments,
Z’ = 1 was used in this simulation. To focus on the intermolecular interactions and to simplify the structure prediction procedure, we did not intend to determine the conformation of the histidine molecule, and used its known conformation found in solid state.
2. Results and Discussion
When examining the data found in ref. [
16] closely, the most information on involvement in intermolecular interactions was obtained for the imidazole moiety of the histidine molecule (
Table 1). Three intermolecular hydrogen-hydrogen interactions were detected; namely, H5···H8’, H5···H7’ and H6···H8’ (for numbering see
Figure 1). Additionally, there were the two above-mentioned nitrogen-hydrogen interactions, H5···N7’ and N5···H7’. The rest of the intermolecular interactions described belong to the interaction between the imidazole moiety and the CH
2 group (H8···H3) and 3 interactions of histidine with water molecules trapped in the crystal lattice (H1···w, H5···w, H8···w).
Generally, for the construction of a 3D network of molecular packing, it is necessary to start with a single molecule of proper geometry. To establish an initial molecule of proper geometry, intramolecular distances can be used. In this work, the original coordinates found in
l-histidine·HCl·H
2O from neutron diffraction are used for simplification [
19]. Subsequently, the geometry optimization of the crystal structure was performed with the CASTEP program [
20].
The obtained molecular geometry was used for the creation of a molecular pair in the mutual arrangement meeting the geometric restrictions based on available intermolecular distances. The initial molecule, labeled as molecule A in the following text, is fixed in space. The second molecule (molecule B) is then moved into proximity with molecule A and arranged in 3D space, meeting the requirements given by the intermolecular interactions listed above. To define the position of a single atom of molecule B with respect to molecule A, three independent distances between this atom and three atoms in molecule A are necessary. However, a set of three distances leads to two possible solutions related by a plane of symmetry defined by the three atoms in molecule A. When defining the position of a whole molecule B, a second set of three distances is required. For the case of
l-histidine·HCl·H
2O, three different distances were found, e.g., for hydrogen H5, namely d(H5···H7’) = 3.15 Å, d(H5···H8’) = 3.25 Å and d(H5···N7’) = 3.44 Å (
Table 1). Unfortunately, the three spheres defined by this distance do not intersect in a single point. A similar situation was found for atom H8, for which three interactions were also given, namely d(H3···H8’) = 2.90 Å, d(H5···H8’) = 3.25 Å and d(H6···H8’) = 3.03 Å (
Table 1). Obviously, the three distances originate from interactions with at least two different molecules in the crystal lattice. Such a possibility always has to be taken into account. Overall, a specific position of H5 or H8 toward molecule A cannot be defined with certainty using the available data.
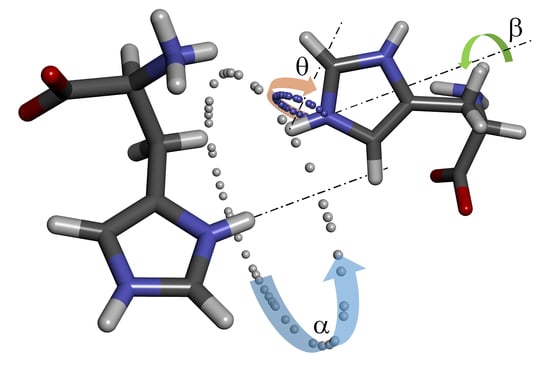
To narrow the space where molecule B can be located and to define its proper orientation, it was necessary to find other interactions that could be attributed to a single molecule. Useful geometric restrictions were found in the imidazole moiety, namely a combination of three interatomic distances between four nuclei N5–H5···H7’–N7’. The imidazole moiety is very convenient, as its geometry can be easily optimized, and the following steps can be also performed just with the imidazole fragment of the molecule. The hydrogen atom H7’ of molecule B has to be placed at a distance of 3.15 Å from hydrogen H5. This constraint reduced the possible space to a sphere with a radius of 3.15 Å centered on H5. Simultaneously, the second constraint keeps hydrogen H7’ at a distance of 3.44 Å from nitrogen N5, reducing the available space from a sphere to a circle (
Figure 2). Each location on such a ring can be characterized by angle α defined as H7
0’–H5–H7’ angle, where H7
0’ is an arbitrarily chosen initial position of H7’.
Having H7’ fixed at the circle around molecule A, nitrogen N7’ has to be placed at a distance of 3.84 Å from hydrogen H5. This constraint locates N7’ on a smaller circle around any chosen position of H7’. The graphical presentation of the possible location of N7’ resembles precession around the imaginary axis H5···H7’. The chosen orientation of molecule B can then be described by precession angle θ (N5–H5–H7’–N7’). Obviously, molecule B can also freely rotate around the H7’–N7’ bond. This rotation defines the last torsion angle (β, H5–H7’–N7’–C6), which is necessary for a complete description of the position and orientation of molecule B towards original molecule A under the given geometric constraints (
Figure 3).
Inspection of the 3D space defined by the three angles defined using a 5-degree step for each angle provides over 370,000 possible arrangements of two histidine molecules. Naturally, a significant part of these solutions can be immediately omitted due to mutual intersection of van der Waals radii of some atoms. On the other hand, some of the solutions evince the approaching of hydrogen H8’ to hydrogen H6 to a distance of 3.03 Å, which was reported as another observable intermolecular interaction,
vide supra. The additional condition of d(H6···H8’) = 3.03 Å reduces the 3D space significantly to the surface of the spherical shape depicted in
Figure 4. The number of possible arrangements using the 5° grid drops to several thousand.
By systematically selecting one particular arrangement of the two histidine molecules after another and using it as a basic repetitive motif, it is possible to start the formation of larger conglomerates. One of the consequences of the symmetrical arrangement of molecules in the crystal lattice is that every molecule must have the same surroundings as the others, considering one independent molecule in the asymmetric unit. Therefore, multiplying a pair of histidine molecules and placing molecule A into the position of molecule B generates another molecule (molecule C). This step can be repeated to generate a molecular chain of an arbitrary length. The mutual arrangement of molecule B to molecule A is in this way transposed to the spacing of molecule C to molecule B and so on (
Figure 5).
As a consequence of crystal lattice periodicity, certain arrangements provide linear chains with identical distances between equivalent atoms in molecules A and C, while the other solutions provide helical structures. The helical arrangement can be acceptable but only for an exact 2-, 3-, 4- or 6-fold axis, as only such axes can be found in real crystal systems. The additional condition of chain linearity or defined helicity further reduces the scope of possible solutions. The number of possible solutions dropped to six using 1-degree incrementation of the three rotation angles. One particular solution (
Figure 6a) places hydrogen H8’’ (molecule C) at a distance of 3.25 Å from hydrogen H5 (molecule A), in accordance with the list of found intermolecular interactions (
Table 1). For examples of molecular chains obtained, see
Figure 6b,c.
In summary, the arrangement of histidine molecules within a final chain was accomplished using six constraints. Three distances in the N5–H5···H7’–N7’ system, interactions H5···H8’ and H6···H8’, and the condition of chain linearity provided a unique solution. The definition of the basic repetitive motif is an essential step for the subsequent construction of the 3D molecular packing. Such a process was not attempted here; therefore, the following paragraphs serve just as a tentative indication of how it could be done.
The chain formed contains translation information on histidine molecules in one dimension. This translation might coincide with one of the cell parameters. To reveal the periodicity in the second and third dimension, the chains have to be assembled into sheets (2D) and the sheets into the final 3D structure. For this purpose, the rest of the available intermolecular interactions can be used in order to control the mutual assembly of molecular chains. The so-far-unused intermolecular interaction between H3···H8’ can be utilized for arrangement of two adjacent chains. A sphere with a radius of 2.90 Å made around each H8 atom in the chain defines the possible location of H3 atoms from the second chain (
Figure 7a). Van der Waals presentation reveals a relatively narrow range in the space where the second chain can be placed (
Figure 7b).
The axes of both chains have to be parallel in order to fulfill d(H3···H8’) = 2.90 Å for each pair of atoms. However, there are two possible arrangements generated by rotation of the second chain axis by 180 degrees (
Figure 8). Furthermore, when the second chain is located at d(H3···H8’) = 2.90 Å, it can be tilted around the axis defined by H3 atoms in the chain arbitrarily until the violation of van der Waals radii is reached. The position of the second chain is defined by d(H3···H8’) distance and angle γ of the inclination. Furthermore, some space has to be left for the positioning of the water molecule. Unfortunately, its location in the crystal lattice can be determined only approximately, as signals of both its hydrogen atoms coincide in the published spectra. However, two of three intermolecular interactions attributed to the water molecule indicate its probable location. The intersection of spheres defined by d(H5···Hw) = 2.47 Å and d(H8···Hw) = 3.09 Å localizes possible placement of the water molecule among histidine molecules.
For the 2D-sheet generation, the chain pair is multiplied, and chain A is placed into the position of chain B, generating another chain, similar to the manner described for chain generation from a pair of histidine molecules. Only chains of certain mutual orientation can provide a linear 2D-sheet in which corresponding atoms in chain A and C are equidistant. The 2D-sheet formed contains translation information in the second dimension (
Figure 9) which might coincide with one of the cell parameters. Perpendicular arrangements of histidine molecules in the sheet (chains A and C, see horizontal lines in
Figure 9) to histidine molecules within a single chain (molecules A and C) should be tested preferentially, as perpendicular arrangements can provide perpendicular cell parameters in the final step.
An assembly of 2D-sheets into a 3D structure represents the final step of crystal lattice reconstruction. The multiplied sheets are brought closer. Individual sheets mutually interlock well and minimize voids in between. The only criterion is the possible collision of van der Waals radii. There are no distance restraints left, which might be used for the mutual positioning of neighboring 2D-sheets. However, the sheets should be positioned so that the volume of empty space in the crystal is minimized (
Figure 10).
Finally, the approximate position of the water molecule in the crystal packing can be determined. However, it cannot be located precisely, as its hydrogen atoms are not spatially resolved in the available NMR data. The accurate location and orientation of the water molecule has to be examined ex post, taking into account possible involvement in hydrogen-bond networking. On the other hand, the location of the chloride anion cannot be determined at all, as there is no information of chloride atom involvement in the molecular packing. Therefore, the remaining voids in the crystal structure and possible hydrogen-bonding interactions have to be examined for its location.
Once the molecular packing is established, it is possible to derive crystal cell parameters. This information is hidden in the periodicity of a single histidine molecule, of each of its atoms. Selecting one particular atom in the histidine molecule and deleting the rest of them, the information on translation becomes obvious (
Figure 11). Then cell parameters are easily determined, and the structure can be classified into the corresponding system, in this case an orthogonal crystal system identical with the system found in ref. [
16].
For this particular solution, parameter
a is found in the 2D-sheet between chain A and C (see
Figure 9). Parameter
b is identified as the distance between 2D-sheet A and B (see
Figure 10). Finally, parameter
c emerges as the distance between histidine molecules A and C within a single molecular chain (see
Figure 5). Four histidine molecules were found in the crystal unit cell. One molecule is situated at the origin of the orthogonal system while the remaining positions correspond to the symmetry operations characteristic for the given space group. The deduction of possible space groups is then rather straightforward. There are 59 space groups available for the orthorhombic crystal system. The requirements for the space group are two: a primitive cell based on the resulting molecular packing (
Figure 11b) and no mirror planes, as histidine is a chiral compound and only its
l-form is present. Therefore, the number of possible space groups is reduced to 4, namely P222, P222
1, P2
12
12 and P2
12
12
1. Based on symmetry operations, each space group generates different positions for four histidine molecules in the unit cell. The only match of positions generated by symmetry operations with the positions found in the reconstructed crystal structure was found for space group P2
12
12
1 providing x, y, z, −x + 1/2, −y, z + 1/2, x + 1/2, −y + 1/2, −z and –x + 1/1, y + 1/2, −z + 1/2 sites.
The found crystallographic parameters can then be confronted with X-ray powder diffraction data for verification of the crystal model proposed. When a significant match is found, the crystal structure can be further refined by Rietveld refinement. Alternatively, the crystal’s structure can be optimized by quantum-chemical methods and NMR parameters can be calculated, which may serve for comparison [
21] with experimental data.