The Length of Hydrophobic Chain in Amphiphilic Polypeptides Regulates the Efficiency of Gene Delivery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Characterization
2.3. Drying of Solvents
2.4. Synthesis of l-Lysine(CBZ)-NCAs Monomer
2.5. Synthesis of l-Leucine-NCAs Monomer
2.6. Synthesis of Poly(l-Lysine(CBZ)50-Block-Poly(l-Leucine)s
2.7. Deprotection to Produce Poly(CBZ-l-Lysine)50-Block-Poly(l-Leucine)s
2.8. Assembly of Poly(l-Lysine)-Block-Poly(l-Leucine) with pDNA
2.9. Investigation of the Biophysical Properties of Polyplexes
2.10. Toxicity Assessment by MTT
2.11. Confocal Laser Scanning Microscopy (CLSM)
2.12. Gene Transfection
2.13. Statistical Analysis
3. Results and Discussion
3.1. Synthesis of NCA Monomers
3.2. Controllable Polymerization
3.3. Deprotection to Produce Polypeptides
3.4. Analysis of Biophysical Properties
3.5. Intracellular Tracking
3.6. Cytotoxicity Evaluation
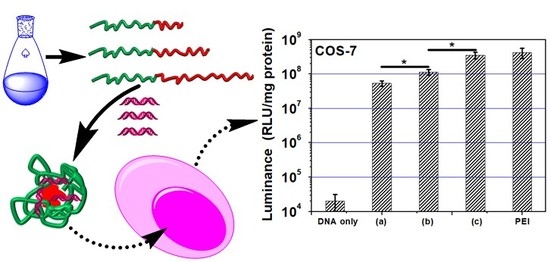
3.7. Gene Transfection
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chen, M.; Wu, J.; Zhou, L.; Jin, C.; Tu, C.; Zhu, B.; Wu, F.; Zhu, Q.; Zhu, X.; Yan, D. Hyperbranched glycoconjugated polymer from natural small molecule kanamycin as a safe and efficient gene vector. Polym. Chem. 2011, 2, 2674–2682. [Google Scholar] [CrossRef]
- Chen, M.; Hu, M.; Wang, D.; Wang, G.; Zhu, X.; Yan, D.; Sun, J. Multifunctional hyperbranched glycoconjugated polymers based on natural aminoglycosides. Bioconj. Chem. 2012, 23, 1189–1199. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhu, X.; Yan, D. A controlled release system for simultaneous promotion of gene transfection and antitumor effects. RSC Adv. 2014, 4, 64596–64600. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, Z.; Zhu, X.; Chen, M. A smart gene delivery platform: Cationic oligomer. Eur. J. Pharm. Sci. 2017, 105, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Kanasty, R.; Dorkin, J.R.; Vegas, A.; Anderson, D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013, 12, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Pai, S.I.; Lin, Y.Y.; Macaes, B.; Meneshian, A.; Hung, C.F.; Wu, T.C. Prospects of RNA interference therapy for cancer. Gene Ther. 2006, 13, 464–477. [Google Scholar] [CrossRef] [PubMed]
- Lorenzer, C.; Dirin, M.; Winkler, A.M.; Baumann, V.; Winkler, J. Going beyond the liver: Progress and challenges of targeted delivery of siRNA therapeutics. J. Controll. Release 2015, 203, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Synatschke, C.V.; Schallon, A.; Jérôme, V.; Freitag, R.; Müller, A.H. Influence of polymer architecture and molecular weight of poly(2-(dimethylamino)ethyl methacrylate) polycations on transfection efficiency and cell viability in gene delivery. Biomacromolecules 2011, 12, 4247–4255. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Jiang, X.; Pan, D.; Mao, H.Q. Charge density and molecular weight of polyphosphoramidate gene carrier are key parameters influencing its DNA compaction ability and transfection efficiency. Biomacromolecules 2010, 11, 3432–3439. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, Z.; Zhou, C.; Jiao, Y. Hydrophobic modifications of cationic polymers for gene delivery. Prog. Polym. Sci. 2010, 35, 1144–1162. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, J.; Xu, D. Cell-penetrating peptides as noninvasive transmembrane vectors for the development of novel multifunctional drug-delivery systems. J. Controll. Release 2016, 229, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, J.; Kanvinde, S.; Lin, Z.; Hazeldine, S.; Singh, R.K.; Oupický, D. Self-immolative polycations as gene delivery vectors and prodrugs targeting polyamine metabolism in cancer. Mol. Pharm. 2015, 12, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.G. Degradation of Synthetic Polypeptides. III. Degradation of Poly-α,l-lysine by Proteolytic Enzymes in 0.20 M Sodium Chloride. J. Am. Chem. Soc. 1964, 86, 3918–3922. [Google Scholar] [CrossRef]
- Putnam, D.; Gentry, C.A.; Pack, D.W.; Langer, R. Polymer-based gene delivery with low cytotoxicity by a unique balance of side-chain termini. Proc. Natl. Acad. Sci. USA 2001, 98, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Midoux, P.; Monsigny, M. Efficient gene transfer by histidylated polylysine/pDNA complexes. Bioconj. Chem. 1999, 10, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Oulad-Abdelghani, M.; Zelkin, A.N.; Wang, Y.; Haîkel, Y.; Mainard, D.; Voegel, J.C.; Caruso, F.; Benkirane-Jessel, N. Poly(l-lysine) nanostructured particles for gene delivery and hormone stimulation. Biomaterials 2010, 31, 1699–1706. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.C.; Kim, S.H.; Jeong, J.H.; Park, T.G. Folate receptor-mediated gene delivery using folate-poly(ethylene glycol)-poly(l-lysine) conjugate. Macromol. Biosci. 2005, 5, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zheng, N.; Song, Z.; Yin, L.; Cheng, J. The effect of side-chain functionality and hydrophobicity on the gene delivery capabilities of cationic helical polypeptides. Biomaterials 2014, 35, 3443–3454. [Google Scholar] [CrossRef] [PubMed]
- Husale, S.; Grange, W.; Karle, M.; Bürgi, S.; Hegner, M. Interaction of cationic surfactants with DNA: A single-molecule study. Nucleic Acids Res. 2008, 36, 1443–1449. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Qiu, M.; Deng, C.; Meng, F.; Zhang, J.; Cheng, R.; Zhong, Z. pH-Responsive chimaeric pepsomes based on asymmetric poly(ethylene glycol)‑b‑poly(l‑leucine)‑b‑poly(l‑glutamic acid) triblock copolymer for efficient loading and active intracellular delivery of Doxorubicin hydrochloride. Biomacromolecules 2015, 16, 1322–1330. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Gao, N.; Wang, Y.; Yi, H.; Fang, S.; Ma, Y.; Cai, L. Self-Assembled Cationic Micelles Based on PEG-PLL-PLLeu Hybrid Polypeptides as Highly Effective Gene Vectors. Biomacromolecules 2012, 13, 3795–3804. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.X.; Merianos, H.J.; Brunger, A.T.; Engelman, D.M. Polar residues drive association of polyleucine transmembrane helices. Proc. Natl. Acad. Sci. USA 2001, 98, 2250–2255. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Lewis, R.N.A.H.; Hodges, R.S.; McElhaney, R.N. Effect of Variations in the structure of a polyleucine-based α-helical transmembrane peptide on its interaction with phosphatidylglycerol bilayers. Biochemistry 2004, 43, 3679–3687. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zhuang, X.; Tang, Z.; Tian, H.; Chen, X. Stimuli-sensitive synthetic polypeptide-based materials for drug and gene delivery. Adv. Healthc. Mater. 2012, 1, 48–78. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Fu, X.; Fu, W.; Li, Z. Biodegradable stimuli-responsive polypeptide materials prepared by ring opening polymerization. Chem. Soc. Rev. 2015, 44, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Erbacher, P.; Roche, A.C.; Monsigny, M.; Midoux, P. The reduction of the positive charges of polylysine by partial gluconoylation increases the transfection efficiency of polylysiner DNA complexes. Biochim. Biophys. Acta 1997, 1324, 27–36. [Google Scholar] [CrossRef]
- Tardy, A.; Nicolas, J.; Gigmes, D.; Lefay, C.; Guillaneuf, Y. Radical ring-opening polymerization: Scope, limitations, and application to (bio)degradable materials. Chem. Rev. 2017, 117, 1319–1406. [Google Scholar] [CrossRef] [PubMed]
- Zetterlund, P.B.; Thickett, S.C.; Perrier, S.; Bourgeat-Lami, E.; Lansalot, M. Controlled/Living radical polymerization in dispersed systems: An update. Chem. Rev. 2015, 115, 9745–9800. [Google Scholar] [CrossRef] [PubMed]
- Deming, T.J. Facile synthesis of block copolypeptides of defined architecture. Nature 1997, 390, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Cheng, J. Hexamethyldisilazane-Mediated Controlled Polymerization of α-Amino Acid N-Carboxyanhydrides. J. Am. Chem. Soc. 2007, 129, 14114–14115. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Deming, T.J. Synthesis of polypeptides by ring-opening polymerization of α-amino acid N-carboxyanhydrides. Top. Curr. Chem. 2012, 310, 1–26. [Google Scholar] [PubMed]
- Jones, C.H.; Chen, C.K.; Ravikrishnan, A.; Rane, S.; Pfeifer, B.A. Overcoming Nonviral Gene Delivery Barriers: Perspective and Future. Mol. Pharm. 2013, 10, 4082–4098. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.B.; Chen, M.; Chen, C.K.; Pfeifer, B.A.; Jones, C.H. Overcoming Gene-Delivery Hurdles: Physiological Considerations for Nonviral Vectors. Trends Biotechnol. 2016, 34, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Hartono, S.B.; Gu, W.; Kleitz, F.; Liu, J.; He, L.; Middelberg, A.P.J.; Yu, C.; Lu, G.Q.; Qiao, S.Z. Poly-l-lysine functionalized large pore cubic mesostructured silica nanoparticles as biocompatible carriers for gene delivery. ACS Nano 2012, 6, 2104–2117. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.N.; van der Aa, M.A.E.M.; Macaraeg, N.; Lee, A.P.; Szoka, F.C., Jr. Quantification of plasmid DNA copies in the nucleus after lipoplex and polyplex transfection. J. Controll. Release 2009, 135, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Mahato, R.I. Lipid and polymeric carrier-mediated nucleic acid delivery. Expert Opin. Drug Deliv. 2010, 7, 1209–1226. [Google Scholar] [CrossRef] [PubMed]
- Ainalem, M.L.; Bartles, A.; Muck, J.; Dias, R.S.; Carnerup, A.M.; Zink, D.; Nylander, T. DNA compaction induced by a cationic polymer or surfactant impact gene expression and DNA degradation. PLoS ONE 2014, 9, e92692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Jensen, S.P.; Hill, M.R.; Moore, G.; He, Z.; Sumerlin, B.S. Synthesis of amphiphilic polysuccinimide star copolymers for responsive delivery in plants. Chem. Commun. 2015, 51, 9694–9697. [Google Scholar] [CrossRef] [PubMed]
- Fissan, H.; Ristig, S.; Kaminski, H.; Asbach, C.; Epple, M. Comparison of different characterization methods for nanoparticle dispersions before and after aerosolization. Anal. Methods 2014, 6, 7324–7334. [Google Scholar] [CrossRef]
- Patel, M.M.; Anchordoquy, T.J. Contribution of Hydrophobicity to Thermodynamics of Ligand-DNA Binding and DNA Collapse. Biophys. J. 2005, 88, 2089–2103. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.C.; Zhang, Q.; Xia, Y. The effect of sedimentation and diffusion on cellular uptake of gold nanoparticles. Nat. Nanotechnol. 2011, 6, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, U.; Maeda, H.; Jain, R.K.; Sevick-Muraca, E.M.; Zamboni, W.; Farokhzad, O.C.; Barry, S.T.; Gabizon, A.; Grodzinski, P.; Blakey, D.C. Challenges and key considerations of the enhanced permeability and retention effect for nanomedicine drug delivery in oncology. Cancer Res. 2013, 73, 2412–2417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, J.; Lykotrafitis, G.; Bao, G.; Suresh, S. Size-dependent endocytosis of nanoparticles. Adv. Mater. 2009, 21, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kröger, M.; Liu, W.K. Endocytosis of PEGylated nanoparticles accompanied by structural and free energy changes of the grafted polyethylene glycol. Biomaterials 2014, 35, 8467–8478. [Google Scholar] [CrossRef] [PubMed]
- Vandenbroucke, R.E.; Lucas, B.; Demeester, J.; De Smedt, S.C.; Sanders, N.N. Nuclear accumulation of plasmid DNA can be enhanced by non-selective gating of the nuclear pore. Nucleic Acids Res. 2007, 35, e86. [Google Scholar] [CrossRef] [PubMed]
- Peetla, C.; Stine, A.; Labhasetwar, V. Biophysical interactions with model lipid membranes: Applications in drug discovery and drug delivery. Mol. Pharm. 2009, 6, 1264–1276. [Google Scholar] [CrossRef] [PubMed]
- Ashley, C.E.; Carnes, E.C.; Phillips, G.K.; Padilla, D.; Durfee, P.N.; Brown, P.A.; Hanna, T.N.; Liu, J.; Phillips, B.; Carter, M.B.; et al. The targeted delivery of multicomponent cargos to cancer cells by nanoporous particle-supported lipid bilayers. Nat. Mater. 2011, 10, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Nowak, A.P.; Breedveld, V.; Pine, D.J.; Deming, T.J. Unusual salt stability in highly charged diblock co-polypeptide hydrogels. J. Am. Chem. Soc. 2003, 125, 15666–15670. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Song, Z.; Yin, L.; Zheng, N.; Tang, H.; Lu, H.; Gabrielson, N.P.; Lin, Y.; Kim, K.; Cheng, J. Ionic α-helical polypeptides toward nonviral gene delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2015, 7, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Yin, L.; Song, Z.; Ma, L.; Tang, H.; Gabrielson, N.P.; Lu, H.; Cheng, J. Maximizing gene delivery efficiencies of cationic helical polypeptides via balanced membrane penetration and cellular targeting. Biomaterials 2014, 35, 1302–1314. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhou, L.; Zhou, Y.; Li, G.; Zhu, X.; Gu, H.; Jiang, X.; Li, H.; Wu, J.; He, L.; et al. Synthesis and gene delivery of poly(amido amine)s with different branched architecture. Biomacromolecules 2010, 11, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Wang, H.; Shao, N.; Wang, M.; Wang, X.; Cheng, Y. Surface-engineered dendrimers with a diaminododecane core achieve efficient gene transfection and low cytotoxicity. Bioconj. Chem. 2014, 25, 342–350. [Google Scholar] [CrossRef] [PubMed]








| Entry | Mn. Theory (×103 g/mol) | Mn a (×103 g/mol) | Mw/Mn | Conv. b | The Final Products | The Proportion of Hydrophobic Domains |
|---|---|---|---|---|---|---|
| Poly(l-lysine(CBZ))50 | 13.1 | 13.2 | 1.10 | >99% | N/A | N/A |
| Poly(l-lysine(CBZ))50-block-Poly(l-leucine)10 | 14.2 | 14.4 | 1.14 | >99% | Poly(l-lysine)50-block-Poly(l-leucine)10 | 15.1% |
| Poly(l-lysine(CBZ))50-block-Poly(l-leucine)15 | 14.7 | 14.8 | 1.27 | >99% | Poly(l-lysine)50-block-Poly(l-leucine)15 | 20.9% |
| Poly(l-lysine(CBZ))50-block-Poly(l-leucine)25 | 15.9 | 15.8 | 1.3 | >99% | Poly(l-lysine)50-block-Poly(l-leucine)25 | 30.6% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zhou, Z.; Chen, M. The Length of Hydrophobic Chain in Amphiphilic Polypeptides Regulates the Efficiency of Gene Delivery. Polymers 2018, 10, 379. https://0-doi-org.brum.beds.ac.uk/10.3390/polym10040379
Zhang Y, Zhou Z, Chen M. The Length of Hydrophobic Chain in Amphiphilic Polypeptides Regulates the Efficiency of Gene Delivery. Polymers. 2018; 10(4):379. https://0-doi-org.brum.beds.ac.uk/10.3390/polym10040379
Chicago/Turabian StyleZhang, Ying, Zhiping Zhou, and Mingsheng Chen. 2018. "The Length of Hydrophobic Chain in Amphiphilic Polypeptides Regulates the Efficiency of Gene Delivery" Polymers 10, no. 4: 379. https://0-doi-org.brum.beds.ac.uk/10.3390/polym10040379