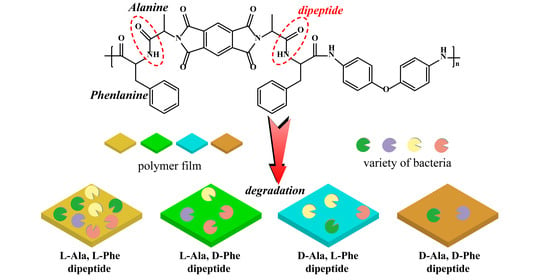
Effect of Homochirality of Dipeptide to Polymers’ Degradation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Characterizations
2.3. Synthesis of Monomer 1–4
2.4. Synthesis of PAIs Containing Dipeptide Structure
2.5. Degradation Experiments of PAI Powders in Phosphate Buffer Solution
2.6. Degradation of PAI Films in Soil
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Toong, D.W.Y.; Toh, H.W.; Ng, J.C.K.; Wong, P.E.H.; Leo, H.L.; Venkatraman, S.; Tan, L.P.; Ang, H.Y.; Huang, Y. Bioresorbable Polymeric Scaffold in Cardiovascular Applications. Int. J. Mol. Sci. 2020, 21, 3444. [Google Scholar] [CrossRef]
- Ruan, C.; Hu, N.; Hu, Y.; Jiang, L.; Cai, Q.; Wang, H.; Pan, H.; Lu, W.W.; Wang, Y. Piperazine-based polyurethane-ureas with controllable degradation as potential bone scaffolds. Polymer 2014, 55, 1020–1027. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Zhu, H.; Shuai, S.; Zhao, C.; Zhou, K.; Ge, W.; Hao, J. Degradation of methoxy-poly (ethylene glycol)-block-poly(α-carboxyl-ε-caprolactone)/magnetite nanocomposites in vitro polymer degradation and stability. Polym. Degrad. Stab. 2020, 177, 109191. [Google Scholar] [CrossRef]
- Yang, Y.; Hu, H.; Chen, L.; Bai, H.; Wang, S.; Xu, J.-F.; Zhang, X. Antibacterial supramolecular polymers constructed via self-sorting: Promoting antibacterial performance and controllable degradation. Mater. Chem. Front. 2019, 3, 806–811. [Google Scholar] [CrossRef]
- Yang, J.-D.; Shu, Y.-T.; Jia, H.-W. Novel controllable degradation behavior and biocompatibility of segmented poly–ε–caprolactone in rats. Polym. Degrad. Stab. 2019, 163, 25–34. [Google Scholar] [CrossRef]
- Park, M.-R.; Cho, C.-S.; Song, S.-C. In vitro and in vivo degradation behaviors of thermosensitive poly(organophosphazene) hydrogels. Polym. Degrad. Stab. 2010, 95, 935–944. [Google Scholar] [CrossRef]
- Froidevaux, V.; Negrell, C.; Caillol, S.; Pascault, J.P.; Boutevin, B. Biobased Amines: From Synthesis to Polymers; Present and Future. Chem. Rev. 2016, 116, 14181–14224. [Google Scholar] [CrossRef]
- Stavila, E.; Loos, K. Synthesis of Polyamides and Their Copolymers via Enzymatic Polymerization. J. Renew. Mater. 2015, 3, 268–280. [Google Scholar] [CrossRef]
- Feng, J.; He, F.; Yang, Z.; Yao, J. Differential study of the biological degradation of polyamide-imides based on the amino acids. Polym. Degrad. Stab. 2016, 129, 231–238. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, L.; Wang, T. Supramolecular Chirality in Self-Assembled Systems. Chem. Rev. 2015, 115, 7304–7397. [Google Scholar] [CrossRef]
- Eschenmoser, A. The search for the chemistry of life’s origin. Tetrahedron 2007, 63, 12821–12844. [Google Scholar] [CrossRef]
- Mallakpour, S.; Khadem, E. A green route for the synthesis of novel optically active poly(amide–imide) nanocomposites containing N-trimellitylimido-l-phenylalanine segments and modified alumina nanoparticles. High Perform. Polym. 2014, 26, 392–400. [Google Scholar] [CrossRef]
- Faghihi, K.; Nourbakhsh, M.; Hajibeygi, M. New optically active poly(amide–imide)s from N-trimellitylimido-l-amino acid and 1,2-bis[4-aminophenoxy]ethane in the main chain: Synthesis and characterization. J. Saudi Chem. Soc. 2014, 18, 783–791. [Google Scholar] [CrossRef] [Green Version]
- Faghihi, K.; Hajibeygi, M.; Shabanian, M. New photosensitive and optically active organo-soluble poly(amide–imide)s from N,N′-(bicyclo[2,2,2]oct-7-ene-tetracarboxylic)-bis-L-amino acids and 1,5-bis(4-aminophenyl)penta-1,4-dien-3-one: Synthesis and characterization. J. Polym. Res. 2009, 17, 379–390. [Google Scholar] [CrossRef]
- Liaw, D.-J.; Chang, F.-C.; Liu, J.-H.; Wang, K.-L.; Faghihi, K.; Huang, S.-H.; Lee, K.-R.; Lai, J.-Y. Novel thermally stable and chiral poly(amide-imide)s bearing from N,N′-(4,4′-diphthaloyl)-bis-l-isoleucine diacid: Synthesis and characterization. Polym. Degrad. Stab. 2007, 92, 323–329. [Google Scholar] [CrossRef]
- Mushtaq, N.; Wang, Q.; Chen, G.; Bashir, B.; Lao, H.; Zhang, Y.; Sidra, L.R.; Fang, X. Synthesis of polyamide-imides with different monomer sequence and effect on transparency and thermal properties. Polymer 2020, 190, 122218. [Google Scholar] [CrossRef]
- Mallakpour, S.; Zadehnazari, A. The production of functionalized multiwall carbon nanotube/amino acid-based poly(amide–imide) composites containing a pendant dopamine moiety. Carbon 2013, 56, 27–37. [Google Scholar] [CrossRef]
- Alborzi, A.R.; Zahmatkesh, S.; Zare, K.; Sadeghi, J. Optically active: Microwave-assisted synthesis and characterization of L-lysine-derived poly (amide-imide)s. Amino Acids 2011, 41, 485–494. [Google Scholar] [CrossRef]
- Li, P.; He, F.; Yang, Z.; Yang, W.; Yao, J. The degradability and thermal properties of chiral polyamide-imides synthesized from several l-amino acids: Side group effects. Polym. Degrad. Stab. 2018, 147, 267–273. [Google Scholar] [CrossRef]
- Dodda, J.M.; Bělský, P. Progress in designing poly(amide imide)s (PAI) in terms of chemical structure, preparation methods and processability. Eur. Polym. J. 2016, 84, 514–537. [Google Scholar] [CrossRef]
- Wu, Q.; Yang, Z.; Yao, J.; Yu, D. Synthesis and biodegradation studies of optically active poly(amide–imide)s based on N,N′-(pyromellitoyl)-bis-l-amino acid. High Perform. Polym. 2015, 28, 34–46. [Google Scholar] [CrossRef]
- Liu, W.; He, F.; Yang, W.; Yang, Z.; Yao, J.; Zhao, H. Study on the Thermal Properties and Enzymatic Degradability of Chiral Polyamide-Imides Films Based on Amino Acids. Appl. Sci. 2019, 9, 578. [Google Scholar] [CrossRef] [Green Version]
- Gan, H.; Tang, K.; Sun, T.; Hirtz, M.; Li, Y.; Chi, L.; Butz, S.; Fuchs, H. Selective adsorption of DNA on chiral surfaces: Supercoiled or relaxed conformation. Angew. Chem. Int. Ed. Engl. 2009, 48, 5282–5286. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Gan, H.; Li, Y.; Chi, L.; Sun, T.; Fuchs, H. Stereoselective Interaction between DNA and Chiral Surfaces. J. Am. Chem. Soc. 2008, 130, 11284–11285. [Google Scholar] [CrossRef] [PubMed]
- Hadavi, E.; de Vries, R.H.W.; Smink, A.M.; de Haan, B.; Leijten, J.; Schwab, L.W.; Karperien, M.; de Vos, P.; Dijkstra, P.J.; van Apeldoorn, A.A. In vitro degradation profiles and in vivo biomaterial-tissue interactions of microwell array delivery devices. J. Biomed. Mater. Res. B Appl. Biomater. 2020. [Google Scholar] [CrossRef]
- Zhao, Q.; Tao, J.; Yam, R.C.M.; Mok, A.C.K.; Li, R.K.Y.; Song, C. Biodegradation behavior of polycaprolactone/rice husk ecocomposites in simulated soil medium. Polym. Degrad. Stab. 2008, 93, 1571–1576. [Google Scholar] [CrossRef]
- Wang, S.; Song, C.; Chen, G.; Guo, T.; Liu, J.; Zhang, B.; Takeuchi, S. Characteristics and biodegradation properties of poly(3-hydroxybutyrate-co-3-hydroxyvalerate)/organophilic montmorillonite (PHBV/OMMT) nanocomposite. Polym. Degrad. Stab. 2005, 87, 69–76. [Google Scholar] [CrossRef]
- Rana, S.K. Blend of high-density polyethylene and a linear low-density polyethylene with compositional-invariant mechanical properties. J. Appl. Polym. Sci. 2002, 83, 2604–2608. [Google Scholar] [CrossRef]
- Newberry, R.W.; Orke, S.J.; Raines, R.T. n-->π* Interactions Are Competitive with Hydrogen Bonds. Org. Lett. 2016, 18, 3614–3617. [Google Scholar] [CrossRef] [Green Version]
- Dill, K.A. Dominant forces in protein folding. Biochemistry 1990, 29, 7133–7155. [Google Scholar] [CrossRef]
- Van Krevelen, D.W.; Hoftyzer, P.J. Properties of Polymers, 3rd ed.; Elsevier Science: Amsterdam, The Netherlands, 1976. [Google Scholar]
- Nelson, M.I. A dynamical systems model of the limiting oxygen index test: II. Retardancy due to char formation and addition of inert fillers. Combust. Theory Model. 2006, 5, 59–83. [Google Scholar] [CrossRef] [Green Version]
- Tamizi, E.; Yang, Y.; Jouyban, A.; Kelso, G.F.; Boysen, R.I.; Hearn, M.T. A capillary electrophoretic-mass spectrometric method for the assessment of octreotide stability under stress conditions. J. Chromatogr. A 2016, 1429, 354–363. [Google Scholar] [CrossRef] [PubMed]





| Polymer | Yield (%) | η (dL/g) 1 | Mw (g/mol) | Formula (Mw) | Elemental Analysis (%) | ||||
|---|---|---|---|---|---|---|---|---|---|
| C | H | N | |||||||
| PAI 1 | 93 | 0.44 | 24530 | −161.11 | (C46H40N6O9)n | Calcd | 67.31 | 4.91 | 10.24 |
| (820.84)n | Found | 67.28 | 4.90 | 10.28 | |||||
| PAI 2 | 90 | 0.40 | 23640 | +105.33 | (C46H40N6O9)n | Calcd | 67.31 | 4.91 | 10.24 |
| (820.84)n | Found | 67.26 | 4.91 | 10.29 | |||||
| PAI 3 | 91 | 0.41 | 23890 | −108.42 | (C46H40N6O9)n | Calcd | 67.31 | 4.91 | 10.24 |
| (820.84)n | Found | 67.21 | 4.98 | 10.27 | |||||
| PAI 4 | 94 | 0.45 | 24820 | +159.65 | (C46H40N6O9)n | Calcd | 67.31 | 4.91 | 10.24 |
| (820.84)n | Found | 67.30 | 4.94 | 10.22 | |||||
| Polymer | Tg (°C) | T5 (°C) 1 | T10 (°C) 2 | Char Yield (%) 3 | LOI 4 |
|---|---|---|---|---|---|
| PAI 1 | 218 | 272 | 316 | 52.35 | 38.44 |
| PAI 2 | 190 | 290 | 312 | 48.48 | 36.89 |
| PAI 3 | 191 | 286 | 315 | 48.20 | 36.78 |
| PAI 4 | 225 | 276 | 318 | 52.26 | 38.40 |
| Polymer | Weight Loss (%) | Molecular Weights Reduction Value (g/mol) |
|---|---|---|
| PAI 1 | 45.63 | 11,560 |
| PAI 2 | 32.08 | 7680 |
| PAI 3 | 26.75 | 6590 |
| PAI 4 | 8.60 | 2170 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; He, F.; Yang, W.; Yao, J. Effect of Homochirality of Dipeptide to Polymers’ Degradation. Polymers 2020, 12, 2164. https://0-doi-org.brum.beds.ac.uk/10.3390/polym12092164
Xu X, He F, Yang W, Yao J. Effect of Homochirality of Dipeptide to Polymers’ Degradation. Polymers. 2020; 12(9):2164. https://0-doi-org.brum.beds.ac.uk/10.3390/polym12092164
Chicago/Turabian StyleXu, Xinqiang, Fuyan He, Wenke Yang, and Jinshui Yao. 2020. "Effect of Homochirality of Dipeptide to Polymers’ Degradation" Polymers 12, no. 9: 2164. https://0-doi-org.brum.beds.ac.uk/10.3390/polym12092164