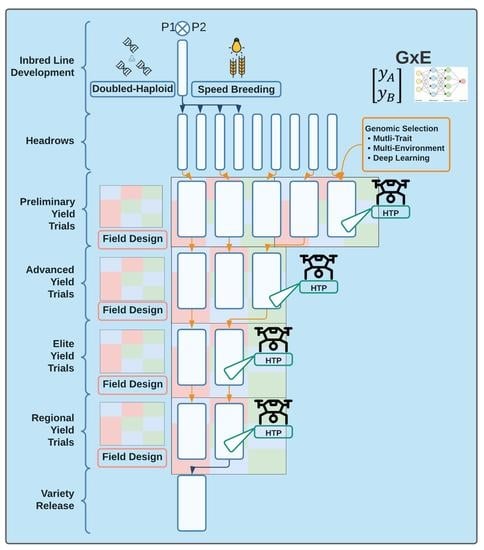
Optimizing Plant Breeding Programs for Genomic Selection
Abstract
:1. Genomic Selection
2. Genetic Gain
3. Breeding Program Optimization
3.1. Speed Breeding and Doubled Haploids
3.2. Training Population Design
3.3. Field Design
4. Leveraging Phenotypic Data
4.1. Multi-Environment Models
4.2. Multi-Trait Models
4.3. Multi-Environment, Multi-Trait Approaches
4.4. Deep Learning
4.5. High-Throughput Phenotyping
5. Genotypic Data and Major Genes
6. Real World Applications
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lorenz, A.J.; Chao, S.; Asoro, F.G.; Heffner, E.L.; Hayashi, T.; Iwata, H.; Smith, K.P.; Sorrells, M.E.; Jannink, J.-L. Genomic Selection in Plant Breeding. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 2011; Volume 110, pp. 77–123. ISBN 978-0-12-385531-2. [Google Scholar]
- Geldermann, H. Investigations on Inheritance of Quantitative Characters in Animals by Gene Markers I. Methods. Theor. Appl. Genet. 1975, 46, 319–330. [Google Scholar] [CrossRef]
- Beckmann, J.S.; Soller, M. Restriction Fragment Length Polymorphisms in Genetic Improvement: Methodologies, Mapping and Costs. Theor. Appl. Genet. 1983, 67, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Botstein, D. Mapping Mendelian Factors Underlying Quantitative Traits Using RFLP Linkage Maps. Genetics 1989, 121, 185–199. [Google Scholar] [CrossRef]
- Lande, R.; Thompson, R. Efficiency of Marker-Assisted Selection in the Improvement of Quantitative Traits. Genetics 1990, 124, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, R. A Model for Marker-Assisted Selection among Single Crosses with Multiple Genetic Markers. Theor. Appl. Genet. 1998, 97, 473–478. [Google Scholar] [CrossRef]
- Bernardo, R. Breeding for Quantitative Traits in Plants, 3rd ed.; Stemma Press: Woodbury, MN, USA, 2020; ISBN 978-0-9720724-3-4. [Google Scholar]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Schaeffer, L.R. Strategy for Applying Genome-Wide Selection in Dairy Cattle. J. Anim. Breed. Genet. 2006, 123, 218–223. [Google Scholar] [CrossRef]
- Massman, J.M.; Jung, H.-J.G.; Bernardo, R. Genomewide Selection versus Marker-Assisted Recurrent Selection to Improve Grain Yield and Stover-Quality Traits for Cellulosic Ethanol in Maize. Crop Sci. 2013, 53, 58. [Google Scholar] [CrossRef] [Green Version]
- Asoro, F.G.; Newell, M.A.; Beavis, W.D.; Scott, M.P.; Tinker, N.A.; Jannink, J.-L. Genomic, Marker-assisted, and Pedigree-BLUP Selection Methods for Β-glucan Concentration in Elite Oat. Crop Sci. 2013, 53, 1894–1906. [Google Scholar] [CrossRef] [Green Version]
- Rutkoski, J.; Singh, R.P.; Huerta-Espino, J.; Bhavani, S.; Poland, J.; Jannink, J.L.; Sorrells, M.E. Genetic Gain from Phenotypic and Genomic Selection for Quantitative Resistance to Stem Rust of Wheat. Plant Genome 2015, 8, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiede, T.; Smith, K.P. Evaluation and Retrospective Optimization of Genomic Selection for Yield and Disease Resistance in Spring Barley. Mol. Breed. 2018, 38, 55. [Google Scholar] [CrossRef]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics. In Harlow Essex UK Longmans Green; Pearson: Essex, UK, 1996; Volume 3. [Google Scholar]
- Cobb, J.N.; Juma, R.U.; Biswas, P.S.; Arbelaez, J.D.; Rutkoski, J.; Atlin, G.; Hagen, T.; Quinn, M.; Ng, E.H. Enhancing the Rate of Genetic Gain in Public-Sector Plant Breeding Programs: Lessons from the Breeder’s Equation. Theor. Appl. Genet. 2019, 132, 627–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaynor, R.C.; Gorjanc, G.; Bentley, A.R.; Ober, E.S.; Howell, P.; Jackson, R.; Mackay, I.J.; Hickey, J.M. A Two-Part Strategy for Using Genomic Selection to Develop Inbred Lines. Crop Sci. 2017, 57, 2372. [Google Scholar] [CrossRef] [Green Version]
- Endelman, J.B.; Atlin, G.N.; Beyene, Y.; Semagn, K.; Zhang, X.; Sorrells, M.E.; Jannink, J.-L. Optimal Design of Preliminary Yield Trials with Genome-Wide Markers. Crop Sci. 2014, 54, 48–59. [Google Scholar] [CrossRef] [Green Version]
- Bassi, F.M.; Bentley, A.R.; Charmet, G.; Ortiz, R.; Crossa, J. Breeding Schemes for the Implementation of Genomic Selection in Wheat (Triticum spp.). Plant Sci. 2016, 242, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Hoefler, R.; González-Barrios, P.; Bhatta, M.; Nunes, J.A.R.; Berro, I.; Nalin, R.S.; Borges, A.; Covarrubias, E.; Diaz-Garcia, L.; Quincke, M.; et al. Do Spatial Designs Outperform Classic Experimental Designs? J. Agric. Biol. Environ. Stat. 2020, 25, 523–552. [Google Scholar] [CrossRef]
- Rutkoski, J.E.; Poland, J.A.; Singh, R.P.; Huerta-Espino, J.; Bhavani, S.; Barbier, H.; Rouse, M.N.; Jannink, J.-L.; Sorrells, M.E. Genomic Selection for Quantitative Adult Plant Stem Rust Resistance in Wheat. Plant Genome 2014, 7, 3. [Google Scholar] [CrossRef] [Green Version]
- Arruda, M.P.; Lipka, A.E.; Brown, P.J.; Krill, A.M.; Thurber, C.; Brown-Guedira, G.; Dong, Y.; Foresman, B.J.; Kolb, F.L. Comparing Genomic Selection and Marker-Assisted Selection for Fusarium Head Blight Resistance in Wheat (Triticum aestivum L.). Mol. Breed. 2016, 36, 84. [Google Scholar] [CrossRef]
- Montesinos-López, O.A.; Montesinos-López, A.; Crossa, J.; Toledo, F.H.; Pérez-Hernández, O.; Eskridge, K.M.; Rutkoski, J. A Genomic Bayesian Multi-Trait and Multi-Environment Model. G3 Genes Genomes Genet. 2016, 6, 2725–2744. [Google Scholar] [CrossRef] [Green Version]
- Sandhu, K.S.; Patil, S.S.; Pumphrey, M.O.; Carter, A.H. Multi-Trait Machine and Deep Learning Models for Genomic Selection Using Spectral Information in a Wheat Breeding Program. Plant Genome 2021, 14, e20119. [Google Scholar] [CrossRef]
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding Crops to Feed 10 Billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.-D.; Asyraf Md Hatta, M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed Breeding Is a Powerful Tool to Accelerate Crop Research and Breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, A.; Hickey, L.T.; Christopher, J.; Rutkoski, J.; Poland, J.; Hayes, B.J. Multivariate Genomic Selection and Potential of Rapid Indirect Selection with Speed Breeding in Spring Wheat. Crop Sci. 2019, 59, 1945. [Google Scholar] [CrossRef] [Green Version]
- Bernardo, R.; Yu, J. Prospects for Genomewide Selection for Quantitative Traits in Maize. Crop Sci. 2007, 47, 1082. [Google Scholar] [CrossRef] [Green Version]
- Heslot, N.; Jannink, J.-L.; Sorrells, M.E. Perspectives for Genomic Selection Applications and Research in Plants. Crop Sci. 2015, 55, 1–12. [Google Scholar] [CrossRef]
- Muleta, K.T.; Bulli, P.; Zhang, Z.; Chen, X.; Pumphrey, M. Unlocking Diversity in Germplasm Collections via Genomic Selection: A Case Study Based on Quantitative Adult Plant Resistance to Stripe Rust in Spring Wheat. Plant Genome 2017, 10, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Belamkar, V.; Guttieri, M.J.; Hussain, W.; Jarquín, D.; El-basyoni, I.; Poland, J.; Lorenz, A.J.; Baenziger, P.S. Genomic Selection in Preliminary Yield Trials in a Winter Wheat Breeding Program. G3 Genes Genomes Genet. 2018, 8, 2735–2747. [Google Scholar] [CrossRef] [Green Version]
- Merrick, L.F.; Carter, A.H. Comparison of Genomic Selection Models for Exploring Predictive Ability of Complex Traits in Breeding Programs. Plant Genome 2021, 14, e20158. [Google Scholar] [CrossRef]
- Merrick, L.F.; Burke, A.B.; Chen, X.; Carter, A.H. Breeding With Major and Minor Genes: Genomic Selection for Quantitative Disease Resistance. Front. Plant Sci. 2021, 12, 1599. [Google Scholar] [CrossRef]
- Lorenz, A.; Nice, L. Training Population Design and Resource Allocation for Genomic Selection in Plant Breeding. In Genomic Selection for Crop Improvement; Springer: Cham, Switzerland, 2017; pp. 7–22. [Google Scholar]
- Asoro, F.G.; Newell, M.A.; Beavis, W.D.; Scott, M.P.; Jannink, J.-L. Accuracy and Training Population Design for Genomic Selection on Quantitative Traits in Elite North American Oats. Plant Genome J. 2011, 4, 132. [Google Scholar] [CrossRef] [Green Version]
- Isidro, J.; Jannink, J.-L.; Akdemir, D.; Poland, J.; Heslot, N.; Sorrells, M.E. Training Set Optimization under Population Structure in Genomic Selection. Theor. Appl. Genet. 2015, 128, 145–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenzana, R.E.; Bernardo, R. Accuracy of Genotypic Value Predictions for Marker-Based Selection in Biparental Plant Populations. Theor. Appl. Genet. 2009, 120, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, R. Genomewide Selection with Minimal Crossing in Self-Pollinated Crops. Crop Sci. 2010, 50, 624. [Google Scholar] [CrossRef]
- Technow, F.; Bürger, A.; Melchinger, A.E. Genomic Prediction of Northern Corn Leaf Blight Resistance in Maize with Combined or Separated Training Sets for Heterotic Groups. G3 Genes Genomes Genet. 2013, 3, 197–203. [Google Scholar] [CrossRef] [Green Version]
- Albrecht, T.; Auinger, H.-J.; Wimmer, V.; Ogutu, J.O.; Knaak, C.; Ouzunova, M.; Piepho, H.-P.; Schön, C.-C. Genome-Based Prediction of Maize Hybrid Performance across Genetic Groups, Testers, Locations, and Years. Theor. Appl. Genet. 2014, 127, 1375–1386. [Google Scholar] [CrossRef]
- Lorenz, A.; Smith, K.P. Adding Genetically Distant Individuals to Training Populations Reduces Genomic Prediction Accuracy in Barley. Crop Sci. 2015, 55, 2657–2667. [Google Scholar] [CrossRef] [Green Version]
- Lozada, D.N.; Godoy, J.V.; Ward, B.P.; Carter, A.H. Genomic Prediction and Indirect Selection for Grain Yield in US Pacific Northwest Winter Wheat Using Spectral Reflectance Indices from High-Throughput Phenotyping. Int. J. Mol. Sci. 2019, 21, 165. [Google Scholar] [CrossRef] [Green Version]
- Lozada, D.N.; Ward, B.P.; Carter, A.H. Gains through Selection for Grain Yield in a Winter Wheat Breeding Program. PLoS ONE 2020, 15, e0221603. [Google Scholar] [CrossRef]
- VanRaden, P.M.; Van Tassell, C.P.; Wiggans, G.R.; Sonstegard, T.S.; Schnabel, R.D.; Taylor, J.F.; Schenkel, F.S. Invited Review: Reliability of Genomic Predictions for North American Holstein Bulls. J. Dairy Sci. 2009, 92, 16–24. [Google Scholar] [CrossRef] [Green Version]
- Mujibi, F.D.N.; Nkrumah, J.D.; Durunna, O.N.; Stothard, P.; Mah, J.; Wang, Z.; Basarab, J.; Plastow, G.; Crews Jr, D.H.; Moore, S.S. Accuracy of Genomic Breeding Values for Residual Feed Intake in Crossbred Beef Cattle. J. Anim. Sci. 2011, 89, 3353–3361. [Google Scholar] [CrossRef] [Green Version]
- Hayes, B.J.; Visscher, P.M.; Goddard, M.E. Increased Accuracy of Artificial Selection by Using the Realized Relationship Matrix. Genet. Res. 2009, 91, 47–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de los Campos, G.; Hickey, J.M.; Pong-Wong, R.; Daetwyler, H.D.; Calus, M.P.L. Whole-Genome Regression and Prediction Methods Applied to Plant and Animal Breeding. Genetics 2013, 193, 327–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norman, A.; Taylor, J.; Edwards, J.; Kuchel, H. Optimising Genomic Selection in Wheat: Effect of Marker Density, Population Size and Population Structure on Prediction Accuracy. G3 Genes Genomes Genet. 2018, 8, 2889–2899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michel, S.; Ametz, C.; Gungor, H.; Akgöl, B.; Epure, D.; Grausgruber, H.; Löschenberger, F.; Buerstmayr, H. Genomic Assisted Selection for Enhancing Line Breeding: Merging Genomic and Phenotypic Selection in Winter Wheat Breeding Programs with Preliminary Yield Trials. Theor. Appl. Genet. 2017, 130, 363–376. [Google Scholar] [CrossRef] [PubMed]
- Berro, I.; Lado, B.; Nalin, R.S.; Quincke, M.; Gutiérrez, L. Training Population Optimization for Genomic Selection. Plant Genome 2019, 12, 190028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gauch Jr, H.G.; Zobel, R.W. Optimal Replication in Selection Experiments. Crop Sci. 1996, 36, 838–843. [Google Scholar] [CrossRef]
- Borges, A.; González-Reymundez, A.; Ernst, O.; Cadenazzi, M.; Terra, J.; Gutiérrez, L. Can Spatial Modeling Substitute for Experimental Design in Agricultural Experiments? Crop Sci. 2019, 59, 44–53. [Google Scholar] [CrossRef] [Green Version]
- Patterson, H.D.; Williams, E.R. A New Class of Resolvable Incomplete Block Designs. Biometrika 1976, 63, 83–92. [Google Scholar] [CrossRef]
- Federer, W.F. Experimental Design; LWW: Cambridge, UK, 1956; Volume 81, ISBN 0038-075X. [Google Scholar]
- Cullis, B.R.; Smith, A.B.; Coombes, N.E. On the Design of Early Generation Variety Trials with Correlated Data. J. Agric. Biol. Environ. Stat. 2006, 11, 381–393. [Google Scholar] [CrossRef]
- Williams, E.; Piepho, H.-P.; Whitaker, D. Augmented P-Rep Designs. Biom. J. 2011, 53, 19–27. [Google Scholar] [CrossRef]
- Gilmour, A.R.; Cullis, B.R.; Verbyla, A.P. Accounting for Natural and Extraneous Variation in the Analysis of Field Experiments. J. Agric. Biol. Environ. Stat. 1997, 2, 269–293. [Google Scholar] [CrossRef] [Green Version]
- González-Barrios, P.; Díaz-García, L.; Gutiérrez, L. Mega-Environmental Design: Using Genotype × Environment Interaction to Optimize Resources for Cultivar Testing. Crop Sci. 2019, 59, 1899–1915. [Google Scholar] [CrossRef] [Green Version]
- Besag, J.; Kempton, R. Statistical Analysis of Field Experiments Using Neighbouring Plots. Biometrics 1986, 231–251. [Google Scholar] [CrossRef]
- Williams, E.R.; John, J.A.; Whitaker, D. Construction of Resolvable Spatial Row–Column Designs. Biometrics 2006, 62, 103–108. [Google Scholar] [CrossRef]
- Ward, B.P.; Brown-Guedira, G.; Tyagi, P.; Kolb, F.L.; Van Sanford, D.A.; Sneller, C.H.; Griffey, C.A. Multienvironment and Multitrait Genomic Selection Models in Unbalanced Early-Generation Wheat Yield Trials. Crop Sci. 2019, 59, 491. [Google Scholar] [CrossRef] [Green Version]
- Federer, W.T. Recovery of Interblock, Intergradient, and Intervariety Information in Incomplete Block and Lattice Rectangle Designed Experiments. Biometrics 1998, 471–481. [Google Scholar] [CrossRef] [Green Version]
- Beeck, C.P.; Cowling, W.A.; Smith, A.B.; Cullis, B.R. Analysis of Yield and Oil from a Series of Canola Breeding Trials. Part I. Fitting Factor Analytic Mixed Models with Pedigree Information. Genome 2010, 53, 992–1001. [Google Scholar] [CrossRef]
- Costa-Neto, G.; Galli, G.; Carvalho, H.F.; Crossa, J.; Fritsche-Neto, R. EnvRtype: A Software to Interplay Enviromics and Quantitative Genomics in Agriculture. G3 Genes Genomes Genet. 2021, 11, jkab040. [Google Scholar] [CrossRef]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; de los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y.; et al. Genomic Selection in Plant Breeding: Methods, Models, and Perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef] [PubMed]
- Burgueño, J.; de los Campos, G.; Weigel, K.; Crossa, J. Genomic Prediction of Breeding Values When Modeling Genotype × Environment Interaction Using Pedigree and Dense Molecular Markers. Crop Sci. 2012, 52, 707–719. [Google Scholar] [CrossRef] [Green Version]
- Heslot, N.; Akdemir, D.; Sorrells, M.E.; Jannink, J.-L. Integrating Environmental Covariates and Crop Modeling into the Genomic Selection Framework to Predict Genotype by Environment Interactions. Theor. Appl. Genet. 2014, 127, 463–480. [Google Scholar] [CrossRef] [PubMed]
- Jarquín, D.; Crossa, J.; Lacaze, X.; Du Cheyron, P.; Daucourt, J.; Lorgeou, J.; Piraux, F.; Guerreiro, L.; Pérez, P.; Calus, M. A Reaction Norm Model for Genomic Selection Using High-Dimensional Genomic and Environmental Data. Theor. Appl. Genet. 2014, 127, 595–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Guo, T.; Mu, Q.; Li, X.; Yu, J. Genomic and Environmental Determinants and Their Interplay Underlying Phenotypic Plasticity. Proc. Natl. Acad. Sci. USA 2018, 115, 6679–6684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lado, B.; Barrios, P.G.; Quincke, M.; Silva, P.; Gutiérrez, L. Modeling Genotype × Environment Interaction for Genomic Selection with Unbalanced Data from a Wheat Breeding Program. Crop Sci. 2016, 56, 2165–2179. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Cruz, M.; Crossa, J.; Bonnett, D.; Dreisigacker, S.; Poland, J.; Jannink, J.-L.; Singh, R.P.; Autrique, E.; de los Campos, G. Increased Prediction Accuracy in Wheat Breeding Trials Using a Marker × Environment Interaction Genomic Selection Model. G3 Genes Genomes Genet. 2015, 5, 569–582. [Google Scholar] [CrossRef] [Green Version]
- Cuevas, J.; Crossa, J.; Soberanis, V.; Pérez-Elizalde, S.; Pérez-Rodríguez, P.; Campos, G.; Montesinos-López, O.A.; Burgueño, J. Genomic Prediction of Genotype x Environment Interaction Kernel Regression Models. Plant Genome 2016, 9, 3. [Google Scholar] [CrossRef] [Green Version]
- Montesinos-López, A.; Montesinos-López, O.A.; Gianola, D.; Crossa, J.; Hernández-Suárez, C.M. Multi-Environment Genomic Prediction of Plant Traits Using Deep Learners with Dense Architecture. G3 Genes Genomes Genet. 2018, 8, 3813–3828. [Google Scholar] [CrossRef] [Green Version]
- Montesinos-López, O.A.; Montesinos-López, A.; Crossa, J.; Gianola, D.; Hernández-Suárez, C.M.; Martín-Vallejo, J. Multi-Trait, Multi-Environment Deep Learning Modeling for Genomic-Enabled Prediction of Plant Traits. G3 Genes Genomes Genet. 2018, 8, 3829–3840. [Google Scholar] [CrossRef] [Green Version]
- Granato, I.; Cuevas, J.; Luna-Vázquez, F.; Crossa, J.; Montesinos-López, O.; Burgueño, J.; Fritsche-Neto, R. BGGE: A New Package for Genomic-Enabled Prediction Incorporating Genotype × Environment Interaction Models. G3 Genes Genomes Genet. 2018, 8, 3039–3047. [Google Scholar] [CrossRef] [Green Version]
- Cuevas, J.; Montesinos-López, O.A.; Martini, J.W.R.; Pérez-Rodríguez, P.; Lillemo, M.; Crossa, J. Approximate Genome-Based Kernel Models for Large Data Sets Including Main Effects and Interactions. Front. Genet. 2020, 11, 1128. [Google Scholar] [CrossRef]
- Montesinos-López, A.; Runcie, D.E.; Ibba, M.I.; Pérez-Rodríguez, P.; Montesinos-López, O.A.; Crespo, L.A.; Bentley, A.R.; Crossa, J. Multi-Trait Genomic-Enabled Prediction Enhances Accuracy in Multi-Year Wheat Breeding Trials. G3 2021, 11, jkab270. [Google Scholar] [CrossRef] [PubMed]
- Ly, D.; Huet, S.; Gauffreteau, A.; Rincent, R.; Touzy, G.; Mini, A.; Jannink, J.-L.; Cormier, F.; Paux, E.; Lafarge, S. Whole-Genome Prediction of Reaction Norms to Environmental Stress in Bread Wheat (Triticum aestivum L.) by Genomic Random Regression. Field Crops Res. 2018, 216, 32–41. [Google Scholar] [CrossRef]
- Millet, E.J.; Kruijer, W.; Coupel-Ledru, A.; Prado, S.A.; Cabrera-Bosquet, L.; Lacube, S.; Charcosset, A.; Welcker, C.; van Eeuwijk, F.; Tardieu, F. Genomic Prediction of Maize Yield across European Environmental Conditions. Nat. Genet. 2019, 51, 952–956. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.; Technow, F.; Messina, C.; Gho, C.; Totir, L.R. Use of Crop Growth Models with Whole-genome Prediction: Application to a Maize Multienvironment Trial. Crop Sci. 2016, 56, 2141–2156. [Google Scholar] [CrossRef] [Green Version]
- Messina, C.D.; Technow, F.; Tang, T.; Totir, R.; Gho, C.; Cooper, M. Leveraging Biological Insight and Environmental Variation to Improve Phenotypic Prediction: Integrating Crop Growth Models (CGM) with Whole Genome Prediction (WGP). Eur. J. Agron. 2018, 100, 151–162. [Google Scholar] [CrossRef]
- Costa-Neto, G.; Fritsche-Neto, R.; Crossa, J. Nonlinear Kernels, Dominance, and Envirotyping Data Increase the Accuracy of Genome-Based Prediction in Multi-Environment Trials. Heredity 2020, 126, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Resende, R.T.; Kuki, K.N.; Correa, T.R.; Zaidan, U.R.; Mota, P.H.S.; Telles, L.A.A.; Gonzales, D.G.; Motoike, S.Y.; Resende, M.D.V.; Leite, H.G. Data-Based Agroecological Zoning of Acrocomia Aculeata: GIS Modeling and Ecophysiological Aspects into a Brazilian Representative Occurrence Area. Ind. Crops Prod. 2020, 154, 112749. [Google Scholar] [CrossRef]
- Bandeira e Sousa, M.; Cuevas, J.; de Oliveira Couto, E.G.; Pérez-Rodríguez, P.; Jarquín, D.; Fritsche-Neto, R.; Burgueño, J.; Crossa, J. Genomic-Enabled Prediction in Maize Using Kernel Models with Genotype × Environment Interaction. G3 Genes Genomes Genet. 2017, 7, 1995–2014. [Google Scholar] [CrossRef] [Green Version]
- Cuevas, J.; Crossa, J.; Montesinos-López, O.A.; Burgueño, J.; Pérez-Rodríguez, P.; de los Campos, G. Bayesian Genomic Prediction with Genotype × Environment Interaction Kernel Models. G3 Genes Genomes Genet. 2017, 7, 41–53. [Google Scholar] [CrossRef] [Green Version]
- Cuevas, J.; Granato, I.; Fritsche-Neto, R.; Montesinos-Lopez, O.A.; Burgueño, J.; Bandeira e Sousa, M.; Crossa, J. Genomic-Enabled Prediction Kernel Models with Random Intercepts for Multi-Environment Trials. G3 Genes Genomes Genet. 2018, 8, 1347–1365. [Google Scholar] [CrossRef] [Green Version]
- Montesinos-López, A.; Montesinos-López, O.A.; Montesinos-López, J.C.; Flores-Cortes, C.A.; de la Rosa, R.; Crossa, J. A Guide for Kernel Generalized Regression Methods for Genomic-Enabled Prediction. Heredity 2021, 126, 577–596. [Google Scholar] [CrossRef]
- Jia, Y.; Jannink, J.-L. Multiple-Trait Genomic Selection Methods Increase Genetic Value Prediction Accuracy. Genetics 2012, 192, 1513–1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, G.; Zhao, F.; Wang, Y.; Zhang, Y.; Du, L.; Su, G. Comparison of Single-Trait and Multiple-Trait Genomic Prediction Models. BMC Genet. 2014, 15, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Zhao, Y.; Rodemann, B.; Plieske, J.; Kollers, S.; Korzun, V.; Ebmeyer, E.; Argillier, O.; Hinze, M.; Ling, J. Potential and Limits to Unravel the Genetic Architecture and Predict the Variation of Fusarium Head Blight Resistance in European Winter Wheat (Triticum aestivum L.). Heredity 2015, 114, 318–326. [Google Scholar] [CrossRef] [Green Version]
- Henderson, C.R.; Quass, R.L. Multiple Trait Evaluation Using Relative’s Record. J. Aim. Sci. 1976, 43, 1188–1197. [Google Scholar]
- Calus, M.P.; Veerkamp, R.F. Accuracy of Multi-Trait Genomic Selection Using Different Methods. Genet. Sel. Evol. 2011, 43, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montesinos-López, O.A.; Montesinos-López, A.; Crossa, J.; Ramírez-Alcaraz, J.M.; Singh, R.; Mondal, S.; Juliana, P. A Singular Value Decomposition Bayesian Multiple-Trait and Multiple-Environment Genomic Model. Heredity 2019, 122, 381–401. [Google Scholar] [CrossRef]
- Bernardo, R. Essentials of Plant Breeding; Stemma Press: Woodbury, MN, USA, 2014; ISBN 978-0-9720724-2-7. [Google Scholar]
- Heffner, E.L.; Sorrells, M.E.; Jannink, J.-L. Genomic Selection for Crop Improvement. Crop Sci. 2009, 49, 1–12. [Google Scholar] [CrossRef]
- Ceron-Rojas, J.J.; Crossa, J.; Arief, V.N.; Basford, K.; Rutkoski, J.; Jarquín, D.; Alvarado, G.; Beyene, Y.; Semagn, K.; DeLacy, I. A Genomic Selection Index Applied to Simulated and Real Data. G3 Genes Genomes Genet. 2015, 5, 2155–2164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, H.S.; Halder, J.; Zhang, J.; Brar, N.K.; Rai, T.S.; Hall, C.; Bernardo, A.; Amand, P.S.; Bai, G.; Olson, E.; et al. Multi-Trait Multi-Environment Genomic Prediction of Agronomic Traits in Advanced Breeding Lines of Winter Wheat. Front. Plant Sci. 2021, 12, 1619. [Google Scholar] [CrossRef]
- Montesinos-López, O.A.; Montesinos-López, A.; Pérez-Rodríguez, P.; Barrón-López, J.A.; Martini, J.W.R.; Fajardo-Flores, S.B.; Gaytan-Lugo, L.S.; Santana-Mancilla, P.C.; Crossa, J. A Review of Deep Learning Applications for Genomic Selection. BMC Genom. 2021, 22, 19. [Google Scholar] [CrossRef] [PubMed]
- Montesinos López, O.A.; Montesinos López, A.; Crossa, J. Fundamentals of Artificial Neural Networks and Deep Learning. In Multivariate Statistical Machine Learning Methods for Genomic Prediction; Montesinos López, O.A., Montesinos López, A., Crossa, J., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 379–425. ISBN 978-3-030-89010-0. [Google Scholar]
- McDowell, R. Genomic Selection with Deep Neural Networks; Iowa State University: Ames, IA, USA, 2016. [Google Scholar]
- Ma, W.; Qiu, Z.; Song, J.; Li, J.; Cheng, Q.; Zhai, J.; Ma, C. A Deep Convolutional Neural Network Approach for Predicting Phenotypes from Genotypes. Planta 2018, 248, 1307–1318. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, K.S.; Lozada, D.N.; Zhang, Z.; Pumphrey, M.O.; Carter, A.H. Deep Learning for Predicting Complex Traits in Spring Wheat Breeding Program. Front. Plant Sci. 2021, 11, 2084. [Google Scholar] [CrossRef] [PubMed]
- Montesinos López, O.A.; Montesinos López, A.; Crossa, J. Artificial Neural Networks and Deep Learning for Genomic Prediction of Continuous Outcomes. In Multivariate Statistical Machine Learning Methods for Genomic Prediction; Montesinos López, O.A., Montesinos López, A., Crossa, J., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 427–476. ISBN 978-3-030-89010-0. [Google Scholar]
- Crain, J.; Mondal, S.; Rutkoski, J.; Singh, R.P.; Poland, J. Combining High-Throughput Phenotyping and Genomic Information to Increase Prediction and Selection Accuracy in Wheat Breeding. Plant Genome 2018, 11, 170043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutkoski, J.; Poland, J.; Mondal, S.; Autrique, E.; Pérez, L.G.; Crossa, J.; Reynolds, M.; Singh, R. Canopy Temperature and Vegetation Indices from High-Throughput Phenotyping Improve Accuracy of Pedigree and Genomic Selection for Grain Yield in Wheat. G3 Genes Genomes Genet. 2016, 6, 2799–2808. [Google Scholar] [CrossRef] [Green Version]
- Krause, M.R.; González-Pérez, L.; Crossa, J.; Pérez-Rodríguez, P.; Montesinos-López, O.; Singh, R.P.; Dreisigacker, S.; Poland, J.; Rutkoski, J.; Sorrells, M.; et al. Hyperspectral Reflectance-Derived Relationship Matrices for Genomic Prediction of Grain Yield in Wheat. G3 Genes Genomes Genet. 2019, 9, 1231–1247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poland, J.A.; Rife, T.W. Genotyping-by-Sequencing for Plant Breeding and Genetics. Plant Genome J. 2012, 5, 92. [Google Scholar] [CrossRef] [Green Version]
- Heffner, E.L.; Lorenz, A.J.; Jannink, J.-L.; Sorrells, M.E. Plant Breeding with Genomic Selection: Gain per Unit Time and Cost. Crop Sci. 2010, 50, 1681. [Google Scholar] [CrossRef]
- Rice, B.; Lipka, A.E. Evaluation of RR-BLUP Genomic Selection Models That Incorporate Peak Genome-Wide Association Study Signals in Maize and Sorghum. Plant Genome 2019, 12, 180052. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Ober, U.; Erbe, M.; Zhang, H.; Gao, N.; He, J.; Li, J.; Simianer, H. Improving the Accuracy of Whole Genome Prediction for Complex Traits Using the Results of Genome Wide Association Studies. PLoS ONE 2014, 9, e93017. [Google Scholar] [CrossRef] [Green Version]
- Spindel, J.E.; Begum, H.; Akdemir, D.; Collard, B.; Redoña, E.; Jannink, J.-L.; McCouch, S. Genome-Wide Prediction Models That Incorporate de Novo GWAS Are a Powerful New Tool for Tropical Rice Improvement. Heredity 2016, 116, 395–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernardo, R. Genomewide Selection When Major Genes Are Known. Crop Sci. 2014, 54, 68. [Google Scholar] [CrossRef]
- Dreisigacker, S.; Crossa, J.; Pérez-Rodríguez, P.; Montesinos-L֯ópez, O.A.; Rosyara, U.; Juliana, P.; Mondal, S.; Crespo-Herrera, L.; Govindan, V.; Singh, R.P. Implementation of Genomic Selection in the CIMMYT Global Wheat Program, Findings from the Past 10 Years. Crop Breed. Genet. Genom. 2021, 3, e210005. [Google Scholar]
- Sneller, C.; Ignacio, C.; Ward, B.; Rutkoski, J.; Mohammadi, M. Using Genomic Selection to Leverage Resources among Breeding Programs: Consortium-Based Breeding. Agronomy 2021, 11, 1555. [Google Scholar] [CrossRef]

| Model | Factor | Description | Software Package (Programming Language) | Reference(s) |
|---|---|---|---|---|
| Two-Step Genomic best linear unbiased prediction (GBLUP) | GE | Environmental and Phenotypic Adjustments made prior to GS using a linear mixed model. | ASREML (R) BGLR (R) | [60] |
| Single-Step GBLUP | GE, MT, MTME | GE GBLUP models using compound symmetry, heterogeneous variance, and factor-analytic unstructured models. | ASREML (R) BGLR (R) | [60] |
| Factor-Analytic (FA) GBLUP | GE | FA GE GBLUP Model | ASREML (R) BGLR (R) | [65] |
| Crop-Growth (CG) covariate GBLUP | GE | CG model derived stress environmental covariates (EC) using the Kronecker product | ASREML (R) BGLR (R) | [66] |
| Reaction-Norm (RN) GBLUP | GE | RN model where the main and interaction effects of markers and environmental covariates are introduced using highly dimensional random variance-covariance structures | BGLR (R) | [67] |
| RN model for phenotypic plasticity (PP) GBLUP | GE | RN GBLUP model for phenotypic plasticity | rrBLUP (R) BGLR (R) | [68] |
| Enviromic-aided (ET) GBLUP | GE | EC GBLUP using Envirotyping | EnvRtype (R) | [63] |
| Genotype-by-Genotype-Environment (GGE) GBLUP | GE | GE based on GGE Mega-environments and additive main-effects and multiplicative interaction (AMMI) using the Kronecker product | rrBLUP (R) BGLR (R) | [69] |
| Marker-Environment Interaction (ME) GBLUP) | GE | ME model that decomposes the marker effects into components common across environments and environment-specific deviations. | BGLR (R) | [70] |
| ME Linear Genome-Based Kernel (GB) GBLUP | GE | ME with the Linear GB kernel | BGLR (R) | [71] |
| ME Gaussian Kernel (GK) GBLUP | GE | ME with the Gaussian GK kernel | BGLR (R) | [71] |
| GB GBLUP | GE, MT, MTME | GE using Kronecker product with the Linear GB GBLUP model | BGLR (R); BMTME (R) | [22,64,72,73] |
| GK GBLUP | GE, MT, MTME | GE using Kronecker product with the Gaussian GK GBLUP model | BGLR (R); BMTME (R) | [22,64,72,73] |
| BGGE GB GBLUP | GE | GE using Hadamard product with the Linear GB GBLUP model | BGGE (R) | [74] |
| BGGE GK GBLUP | GE | GE using Hadamard product with the Gaussian GK GBLUP model | BGGE (R) | [74] |
| Approximate Kernel (AK) RN GBLUP | GE | Sparse Approximate Model using the RN GBLUP model | BGLR (R) | [75] |
| AK GBLUP | GE, MT, MTME | Sparse Approximate Model using the Kronecker product for GB and GK GBLUP along with various other kernels | BGLR (R) | [76] |
| Multi-Layer Perceptron (MLP) | GE, MT, MTME | Deep learning MLP that uses a combination of input, hidden, and output layers using a large number of neurons for building the relationship between the predictors and output that has the ability to incorporate GB and other kernels and use any GE method. | TensorFlow (R and Python) and Keras (R and Python) | [72,73] |
| Model | Description | Software Package (Programming Language) | Reference(s) |
|---|---|---|---|
| Genomic best linear unbiased prediction (GBLUP) | The GBLUP model that uses GRMs for predicting their performance. In addition, has the ability to use single and multi-kernel models combining hyperspectral and genomic marker information | ASReml (R); BGLR (R) | [23,103,104,105] |
| Bayesian | Bayesian models (Bayes A, Bayes B, Bayes C, Bayes Cπ, Bayes D, Bayes Lasso, Bayes Ridge Regression) that use marker effects by assuming a scaled inverted chi-square distribution, scaled t distribution, or double exponential distribution for variance parameters to model marker effects. | BLGR(R); BMTME (R) | [23] |
| Elastic Net (EN) | EN is the intermediate between ridge regression and lasso using an average weight penalty for marker effect estimations. | glmnet (R) | [103] |
| Partial least square regression (PLSR) | PLSR is a dimensional reduction approach that uses latent variables derived from predictors to link with the response variables. | pls (R) | [103] |
| Random Forest (RF) | RF uses a network of the tree with varying number of nodes, resampling, and depth for building the final tree regression for predictions | caret (R); Scikit-learn (Python) | [23] |
| Support-Vector Machine (SVM) | SVM is a non-parametric method that uses kernels functions, and cost functions to model hyperplanes for predictions. | caret (R); Scikit-learn (Python) | [23] |
| Convolutional Neural Network (CNN) | Deep learning CNN that uses convolutional, flattening, pooling, and dense layers for predicting using kernels to reduce the excess predictors from the model. | caret (R); Keras (R and Python); Scikit-learn (Python) | [23] |
| Multi-layer Perceptron (MLP) | Deep learning MLP that uses a combination of input, hidden, and output layers using a large number of neurons for building the relationship between the predictors | caret (R); Keras (R and Python); Scikit-learn (Python) | [23] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merrick, L.F.; Herr, A.W.; Sandhu, K.S.; Lozada, D.N.; Carter, A.H. Optimizing Plant Breeding Programs for Genomic Selection. Agronomy 2022, 12, 714. https://0-doi-org.brum.beds.ac.uk/10.3390/agronomy12030714
Merrick LF, Herr AW, Sandhu KS, Lozada DN, Carter AH. Optimizing Plant Breeding Programs for Genomic Selection. Agronomy. 2022; 12(3):714. https://0-doi-org.brum.beds.ac.uk/10.3390/agronomy12030714
Chicago/Turabian StyleMerrick, Lance F., Andrew W. Herr, Karansher S. Sandhu, Dennis N. Lozada, and Arron H. Carter. 2022. "Optimizing Plant Breeding Programs for Genomic Selection" Agronomy 12, no. 3: 714. https://0-doi-org.brum.beds.ac.uk/10.3390/agronomy12030714