Assessment of Genetic Diversity in Opuntia spp. Portuguese Populations Using SSR Molecular Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and DNA Extraction
2.2. Microsatellite Genotyping
2.3. Data Analyses
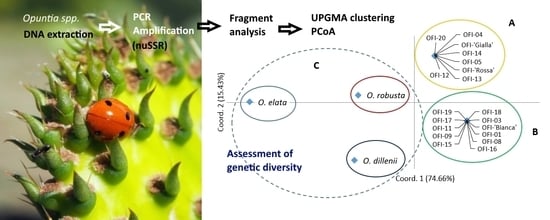
3. Results
3.1. Microsatellite Genotyping
3.2. Genetic Relationships and Diversity Patterns
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Griffith, M.P. The origins of an important cactus crop, Opuntia ficus-indica (Cactaceae): New molecular evidence. Am. J. Bot. 2004, 91, 1915–1921. [Google Scholar] [CrossRef] [PubMed]
- Barbera, G.; Carimi, F.; Inglese, P. Past and present role of the Indian-fig prickly-pear (Opuntia ficus-indica (L.) Miller, Cactaceae) in the Agriculture of Sicily. Econ. Bot. 1992, 46, 10–20. [Google Scholar] [CrossRef]
- Anderson, E. The Cactus Family; Timber Press: Portland, OR, USA, 2001; p. 776. [Google Scholar]
- Nieddu, G.; Chessa, I. Distribution of phenotipic characters within a seedling population from Opuntia ficus-indica (cv. “Gialla”). Acta Hortic. 1997, 438, 37–40. [Google Scholar] [CrossRef]
- Kalia, R.K.; Rai, M.K.; Kalia, S.; Singh, R.; Dhawan, A.K. Microsatellite markers: An overview of the recent progress in plants. Euphytica 2011, 177, 309–334. [Google Scholar] [CrossRef]
- De Cruz, M.; Ramirez, F.; Hernandez, H. DNA isolation and amplification from cacti. Plant Mol. Biol. Report. 1997, 15, 319–325. [Google Scholar] [CrossRef]
- Mondragon-Jacobo, C.; Doudareva, N.; Bordelon, B.P. DNA extraction from several cacti. HortScience 2000, 35, 1124–1126. [Google Scholar]
- Pandey, R.N.; Adams, R.P.; Flournoy, L.E. Inhibition of random amplified polymorphic DNAs (RAPDs) by plant polysaccharides. Plant Mol. Biol. Report. 1996, 14, 17–22. [Google Scholar] [CrossRef]
- Majure, L.C.; Puente, R.; Pinkava, D.J. Miscellaneous chromosome numbers in Opuntieae Dc. (Cactaceae) with a compilation of counts for the group. Haseltonia 2012, 18, 67–78. [Google Scholar] [CrossRef]
- Pinkava, D. On the evolution of the North American Opuntioideae. In Studies in the Opuntioideae (Cactaceae), 1st ed.; Hunt, N., Taylor, D.R., Eds.; DH Books: Hong Kong, China, 2002; pp. 59–98. [Google Scholar]
- Pinkava, D.J.; McLeod, M.G.; McGill, L.A.; Brown, R.C. Chromosome numbers in some cacti of Western North America II. Brittonia 1973, 25, 2–9. [Google Scholar] [CrossRef]
- Segura, S.; Scheinvar, L.; Olalde, G.; Leblanc, O.; Filardo, S.; Muratalla, A.; Gallegos, C.; Flores, C. Genome sizes and ploidy levels in Mexican cactus pear species Opuntia (Tourn.) Mill. series Streptacanthae Britton et Rose, Leucotrichae DC., Heliabravoanae Scheinvar and Robustae Britton et Rose. Genet. Resour. Crop Evol. 2007, 54, 1033–1041. [Google Scholar] [CrossRef]
- Helsen, P.; Verdyck, P.; Tye, A.; Desender, K.; Van Houtte, N.; Van Dongen, S. Primer note: Isolation and characterization of polymorphic microsatellite markers in Galapagos prickly pear (Opuntia) cactus species. Mol. Ecol. Notes 2006, 7, 454–456. [Google Scholar] [CrossRef]
- Erre, P.; Nieddu, G.; Chessa, I. Identification of microsatellite loci in Opuntia spp. and their characterization in cultivars and species. Acta Hortic. 2011, 28, 327–332. [Google Scholar] [CrossRef]
- Helsen, P.; Verdyck, P.; Tye, A.; Van Dongen, S. Low levels of genetic differentiation between Opuntia echios varieties on Santa Cruz (Galapagos). Plant Syst. Evol. 2009, 279, 1–10. [Google Scholar] [CrossRef]
- Caruso, M.; Currò, S.; Las Casas, G.; La Malfa, S.; Gentile, A. Microsatellite markers help to assess genetic diversity among Opuntia ficus-indica cultivated genotypes and their relation with related species. Plant Syst. Evol. 2010, 290, 85–97. [Google Scholar] [CrossRef]
- Samah, S.; De Teodoro Pardo, C.V.; Serrato Cruz, M.A.; Valadez-Moctezuma, E. Genetic diversity, genotype discrimination, and population structure of Mexican Opuntia sp., determined by SSR markers. Plant Mol. Biol. Report. 2016, 34, 146–159. [Google Scholar] [CrossRef]
- López-Vinyallonga, S.; Soriano, I.; Susanna, A.; Montserra, J.M.; Roquet, C.; Garcia-Jacas, N. The polyploid series of the Achillea millefolium aggregate in the Iberian Peninsula investigated using microsatellites. PLoS ONE 2015, 10, e0129861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohlf, F.J. NTSY-SPC Numerical Taxonomy System; Exter Software: New York, NY, USA, 2002. [Google Scholar]
- Mantel, N.A. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar] [PubMed]
- Vekemans, X. AFLP-SURV 1.0; Distributed by the author: Laboratoire de Génétique et Ecologie Végétale; Université Libre de Bruxelles: Brussels, Belgium, 2002. [Google Scholar]
- Peakall, R.; Smouse, P.E. Genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2005, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [PubMed]
- Laurent, E.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar]
- Kirk, H.; Freeland, J.R. Applications and Implications of Neutral versus Non-neutral Markers in Molecular Ecology. Int. J. Mol. Sci. 2011, 12, 3966–3988. [Google Scholar] [CrossRef] [PubMed]
- Freeman, J.S.; Marques, C.M.P.; Carocha, V.; Borralho, N.; Potts, B.M.; Vaillancourt, R.E. Origins and diversity of the Portuguese Landrace of Eucalyptus globulus. Ann. For. Sci. 2007, 64, 639–647. [Google Scholar] [CrossRef]
- Blasco, M.; Angulo, M.J.; Cristina, I. Difference in the invasive potential of Opuntia genus inhabiting the south of Iberian Peninsula. Acta Hortic. 2015, 1067, 67–74. [Google Scholar] [CrossRef]
- Mondragon-Jacobo, C. Verification of the apomictic origin of cactus pear (Opuntia spp. Cactaceae) seedlings of open pollinated and crosses from Central Mexico. JPACD 2001, 4, 49–56. [Google Scholar]
- Erre, P.; Chessa, I.; Nieddu, G.; Jones, P.G. Diversity and spatial distribution of Opuntia spp. in the Mediterranean Basin. J. Arid Environ. 2009, 73, 1058–1066. [Google Scholar] [CrossRef]
- Chessa, G.; Nieddu, I. Investigations on variability in the genus Opuntia as fruit crop for genetic improvement. Acta Hortic. 2002, 575, 345–353. [Google Scholar] [CrossRef]
- Ellegren, H. Polymerase-Chain-Reaction (PCR) analysis of microsatellites: A new approach to studies of genetic relationships in birds. Auk 1992, 109, 886–895. [Google Scholar] [CrossRef]
- Kijas, J.M.H.; Fowler, J.C.S.; Thomas, M.R. An evaluation of sequence tagged microsatellite site markers for genetic analysis within Citrus and related species. Genome 1995, 38, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.E.; Pennington, R.T.; Pennington, T.D.; Hollingsworth, P.M. Rapid diversification of a species-rich genus of neotropical rain forest trees. Science 2001, 293, 2242–2245. [Google Scholar] [CrossRef] [PubMed]
- Reis, C.M.G.; Gazarini, L.C.; Fonseca, T.F.; Ribeiro, M.M. Above-ground biomass estimation of Opuntia ficus-indica (L.) Mill. for forage crop in a Meditterranean environment by using non-destructive methods. Exp. Agric. 2018, 54, 227–242. [Google Scholar] [CrossRef]


| Population | Pulp Colour | Origin | Altitude (m) | Geographic Coordinates | |
|---|---|---|---|---|---|
| Latitude | Longitude | ||||
| OFI-01 a | White | Alcochete | 25 | 38°43′32.14″ N | 8°57′58.22″ W |
| OFI-03 | White | Cascais | 185 | 38°45′23.18″ N | 9°27′38.48″ W |
| OFI-04 | Pale yellow | Portalegre | 372 | 39°16′22.45″ N | 7°26′13.12″ W |
| OFI-05 | Orange | Arronches | 293 | 39°5′21.06″ N | 7°12′7.05″ W |
| OFI-08 | White | Melides | 29 | 38°8′28.91″ N | 8°44′14.28″ W |
| OFI-09 | White | Santo André | 25 | 38°4′38.13″ N | 8°46′38.08″ W |
| OFI-11 | White | Albufeira | 61 | 37°5′23.33″ N | 8°17′27.03″ W |
| OFI-12 | Orange | Cacela-a-Velha | 20 | 37°9′22.50″ N | 7°32′47.98″ W |
| OFI-13 | Orange | Monforte da Beira | 260 | 39°45′8.34″ N | 7°16′54.83″ W |
| OFI-14 | Orange | Idanha-a-Velha | 275 | 39°59′57.30″ N | 7°9′3.51″ W |
| OFI-15 | White | Ponte de Sor | 125 | 39°16′15.45″ N | 8°0′44.72″ W |
| OFI-16 | White | Coruche | 76 | 38°54′40.93″ N | 8°37′17.00″ W |
| OFI-17 | White | Castelo Branco | 402 | 39°48′58.84″ N | 7°29′37.85″ W |
| OFI-18 | White | Reg. Monsaraz | 223 | 38°27′27.04″ N | 7°39′21.77″ W |
| OFI-19 | White | Alvega | 105 | 39°27′15.96″ N | 8°3′51.88″ W |
| OFI-20 | Orange | Madeira | 116 | 32°38′54.18″ N | 16°57′46.38″ W |
| OFI, cv. Bianca | White | Italy | -- | -- | -- |
| OFI, cv. Gialla | Orange | Italy | -- | -- | -- |
| OFI, cv. Rossa | Red | Italy | -- | -- | -- |
| O. robusta | Red | Castelo Branco | 365 | 39°49′17.00″ N | 7°27′41.00″ W |
| O. dillenii | Purple | Lagos | 48 | 37°8′42.24″ N | 8°40′33.42″ W |
| O. elata | Purple | S. J. Pesqueira | 450 | 41°9′5.83″ N | 7°22′5.43″ W |
| Marker | Na | Alleles Per Individual | No. of Rare Alleles * | PIC (Average) | MI |
|---|---|---|---|---|---|
| OA3 | 6 | 2–3 | 3 | 0.635 | 3.808 |
| OA5 | 8 | 2–4 | 4 | 0.738 | 5.903 |
| OA9 | 10 | 2–5 | 5 | 0.495 | 4.948 |
| OA11 | 5 | 2–5 | 1 | 0.705 | 3.524 |
| OA12 | 16 | 3–7 | 7 | 0.882 | 14.106 |
| OA13 | 10 | 2–5 | 4 | 0.812 | 8.120 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reis, C.M.G.; Raimundo, J.; Ribeiro, M.M. Assessment of Genetic Diversity in Opuntia spp. Portuguese Populations Using SSR Molecular Markers. Agronomy 2018, 8, 55. https://0-doi-org.brum.beds.ac.uk/10.3390/agronomy8040055
Reis CMG, Raimundo J, Ribeiro MM. Assessment of Genetic Diversity in Opuntia spp. Portuguese Populations Using SSR Molecular Markers. Agronomy. 2018; 8(4):55. https://0-doi-org.brum.beds.ac.uk/10.3390/agronomy8040055
Chicago/Turabian StyleReis, Carlos M. G., Joana Raimundo, and Maria Margarida Ribeiro. 2018. "Assessment of Genetic Diversity in Opuntia spp. Portuguese Populations Using SSR Molecular Markers" Agronomy 8, no. 4: 55. https://0-doi-org.brum.beds.ac.uk/10.3390/agronomy8040055