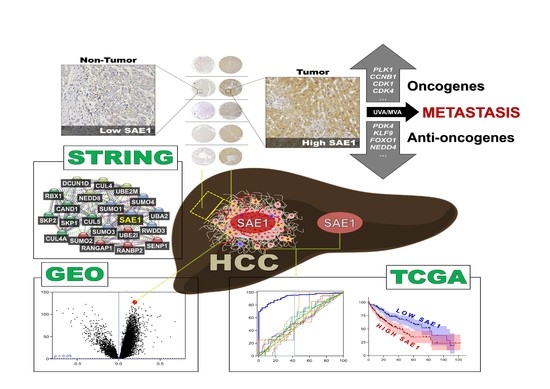
SUMO-Activating Enzyme Subunit 1 (SAE1) Is a Promising Diagnostic Cancer Metabolism Biomarker of Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. HCC Samples and Cohort Characterization
2.2. Data Acquisition and Statistical Analysis of HCC
2.3. Immunohistochemistry
2.4. SAE1 Knockdown Using CRISPR Interference
2.5. Real-Time PCR Reaction
2.6. Western Blot Analysis
2.7. Transwell Matrigel Invasion Assay
2.8. Scratch-Wound Migration Assay
2.9. Statistical Analysis
3. Results
3.1. Gene Expression Profile of SAE1 in Pan-Cancer Cohort
3.2. SAE1 Is Overexpressed in HCC and Associated with Disease Progression
3.3. The Overexpression of SAE1 Is Associated with Metastasis and Poor Prognosis in Patients with HCC
3.4. SAE1 Is a Reliable Diagnostic and Prognostic Biomarker for HCC
3.5. SAE1 Upregulates Oncogenic Effectors of Cell Cycle Progression while Downregulating FOXO1-Associated Tumor Suppressing Signaling
3.6. The Oncogenic Effect of Upregulated SAE1 Is Associated with Dysregulated Cancer Metabolic Signalings
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Fujiwara, N.; Friedman, S.L.; Goossens, N.; Hoshida, Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J. Hepatol. 2018, 68, 526–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jee, B.A.; Choi, J.H.; Rhee, H.; Yoon, S.; Kwon, S.M.; Nahm, J.H.; Yoo, J.E.; Jeon, Y.; Choi, G.H.; Woo, H.G.; et al. Dynamics of Genomic, Epigenomic, and Transcriptomic Aberrations during Stepwise Hepatocarcinogenesis. Cancer Res. 2019, 79, 5500–5512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Losic, B.; Craig, A.J.; Villacorta-Martin, C.; Martins-Filho, S.N.; Akers, N.; Chen, X.; Ahsen, M.E.; von Felden, J.; Labgaa, I.; D’Avola, D.; et al. Intratumoral heterogeneity and clonal evolution in liver cancer. Nat. Commun. 2020, 11, 291. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Zhao, X.; Yang, X.R.; Li, F.Q.; Zhou, X.L.; Wu, K.; Zhang, X.; Sun, Q.-M.; Cao, Y.; Zhu, H.-M.; et al. Circumventing intratumoral heterogeneity to identify potential therapeutic targets in hepatocellular carcinoma. J. Hepatol. 2017, 67, 293–301, Erratum in J. Hepatol. 2017, 67, 1123. [Google Scholar] [CrossRef] [PubMed]
- Kotha, S.; Neong, S.; Patel, K. Serum biomarkers for diagnosis and monitoring viral hepatitis and hepatocellular carcinoma. Expert Rev. Mol. Diagn. 2018, 18, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.S.; Shyu, Y.C.; Turner, R.; Chen, H.Y.; Chen, P.J. Diagnostic performance of alpha-fetoprotein, lens culinaris agglutinin-reactive alpha-fetoprotein, des-gamma carboxyprothrombin, and glypican-3 for the detection of hepatocellular carcinoma: A systematic review and meta-analysis protocol. Syst. Rev. 2013, 2, 37. [Google Scholar] [CrossRef] [Green Version]
- Lou, J.; Zhang, L.; Lv, S.; Zhang, C.; Jiang, S. Biomarkers for Hepatocellular Carcinoma. Biomark Cancer 2017, 9, 1–9. [Google Scholar] [CrossRef]
- Gao, Y.X.; Yang, T.W.; Yin, J.M.; Yang, P.X.; Kou, B.X.; Chai, M.Y.; Liu, X.N.; Chen, D.X. Progress and prospects of biomarkers in primary liver cancer (Review). Int. J. Oncol. 2020, 57, 54–66. [Google Scholar] [CrossRef]
- Geiss-Friedlander, R.; Melchior, F. Concepts in sumoylation: A decade on. Nat. Rev. Mol. Cell Biol. 2007, 8, 947–956. [Google Scholar] [CrossRef]
- Yang, Y.; Liang, Z.; Xia, Z.; Wang, X.; Ma, Y.; Sheng, Z.; Gu, Q.; Shen, G.; Zhou, L.; Zhu, H.; et al. SAE1 promotes human glioma progression through activating AKT SUMOylation-mediated signaling pathways. Cell Commun. Signal. 2019, 17, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Jiang, D.; Xie, X.; He, Y.; Lv, M.; Jiang, X. miR-129-3p inhibits NHEJ pathway by targeting SAE1 and represses gastric cancer progression. Int. J. Clin. Exp. Pathol. 2019, 12, 1539–1547. [Google Scholar] [PubMed]
- Kessler, J.D.; Kahle, K.T.; Sun, T.; Meerbrey, K.L.; Schlabach, M.R.; Schmitt, E.M.; Sninnrt, S.O.; Xu, Q.; Li, M.Z.; Hartman, Z.C.; et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 2012, 335, 348–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.S.; Thorgeirsson, S.S. Genome-Scale profiling of gene expression in hepatocellular carcinoma: Classification, survival prediction, and identification of therapeutic targets. Gastroenterology 2004, 127, S51–S55. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Yang, C.K.; Wei, P.L.; Huynh, T.T.; Whang-Peng, J.; Meng, T.C.; Hsiao, M.; Tzeng, Y.M.; Wu, A.T.; Yen, Y. Ovatodiolide suppresses colon tumorigenesis and prevents polarization of M2 tumor-associated macrophages through YAP oncogenic pathways. J. Hematol. Oncol. 2017, 10, 60. [Google Scholar] [CrossRef] [Green Version]
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar]
- Zubiete-Franco, I.; García-Rodríguez, J.L.; Lopitz-Otsoa, F.; Lopitz-Otsoa, F.; Serrano-Macia, M.; Simon, J.; Fernandez-Tussy, F.; Barbier-Torres, L.; Fernandez-Ramos, D.; Gutierrez-de-Juan, V.; et al. SUMOylation regulates LKB1 localization and its oncogenic activity in liver cancer. EBioMedicine 2019, 40, 406–421. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Tao, X.; Zhang, J.; Wang, P.; Sha, M.; Ma, Y.; Geng, X.; Feng, L.; Shen, Y.; Yu, Y.; et al. Small ubiquitin-related modifier 1 is involved in hepatocellular carcinoma progression via mediating p65 nuclear translocation. Oncotarget 2016, 7, 22206–22218. [Google Scholar] [CrossRef] [Green Version]
- Pichler, A.; Fatouros, C.; Lee, H.; Eisenhardt, N. SUMO conjugation—A mechanistic view. Biomol. Concepts 2017, 8, 13–36. [Google Scholar] [CrossRef]
- Tomasi, M.L.; Tomasi, I.; Ramani, K.; Pascale, R.M.; Xu, J.; Giordano, P.; Mato, J.M.; Lu, S.C. S-Adenosyl methionine regulates ubiquitin-conjugating enzyme 9 protein expression and sumoylation in murine liver and human cancers. Hepatology 2012, 56, 982–993. [Google Scholar] [CrossRef]
- Han, Z.J.; Feng, Y.H.; Gu, B.H.; Li, Y.M.; Chen, H. The post-translational modification, SUMOylation, and cancer (Review). Int. J. Oncol. 2018, 52, 1081–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, R.; Fang, J.; Liu, M.; Jun, A.; Liu, J.; Chen, W.; Li, J.; Ma, G.; Zhang, Z.; Zhang, B.; et al. SUMOylation of the transcription factor ZFHX3 at Lys-2806 requires SAE1, UBC9, and PIAS2 and enhances its stability and function in cell proliferation. J. Biol. Chem. 2020, 295, 6741–6753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Gao, S.; Chen, J.; Lou, W. UBE2I promotes metastasis and correlates with poor prognosis in hepatocellular carcinoma. Cancer Cell Int. 2020, 20, 234. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, J.J.; Pelluru, D.; Lefkimmiatis, K.; Fulciniti, M.; Prabhala, R.H.; Greipp, P.R.; Barlogie, B.; Tai, Y.-T.; Anderson, K.C.; Shaughnessy, J.D.; et al. The sumoylation pathway is dysregulated in multiple myeloma and is associated with adverse patient outcome. Blood 2010, 115, 2827–2834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, Y.S.; Cho, S.J.; Park, J.; Kim, Y.; Choi, Y.J.; Pyo, J.S.; Jang, B.G.; Park, J.-W.; Kim, W.H.; Lee, B.L. Loss of FOXO1 promotes gastric tumour growth and metastasis through upregulation of human epidermal growth factor receptor 2/neu expression. Br. J. Cancer 2015, 113, 1186–1196. [Google Scholar] [CrossRef] [Green Version]
- Hendriks, I.A.; Vertegaal, A.C. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 2016, 17, 581–595. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Parvin, J.D. Small ubiquitin-like modifier (SUMO) isoforms and conjugation-independent function in DNA double-strand break repair pathways. J. Biol. Chem. 2014, 289, 21289–21295. [Google Scholar] [CrossRef] [Green Version]
- Barr, A.R.; Cooper, S.; Heldt, F.S.; Butera, F.; Stoy, H.; Mansfeld, J.; Novák, B.; Bakal, C. DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression. Nat. Commun. 2017, 8, 14728. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.; Tian, L.; Liu, S. Combining large number of weak biomarkers based on AUC. Stat. Med. 2015, 34, 3811–3830. [Google Scholar] [CrossRef] [Green Version]
- Zou, K.H.; Liu, A.; Bandos, A.I.; Ohno-Machado, L.; Rockette, H.E. Statistical Evaluation of Diagnostic Performance; Chapman and Hall/CRC: Boca Raton, FL, USA, 2011; p. 33487. [Google Scholar]






| Clinicopathological Variables | Low SAE1 (n = 25) | High SAE1 (n = 29) | p-Value | ||
|---|---|---|---|---|---|
| Gender (n, %) | |||||
| Male | 7 | 28 | 2 | 6.9 | 0.088 |
| Female | 18 | 72 | 27 | 93.1 | |
| Tumor Stage (n, %) | |||||
| I + II | 18 | 72 | 14 | 48.3 | 0.021 * |
| III + IV | 7 | 28 | 15 | 51.7 | |
| Metastasis (n, %) | |||||
| M0 | 18 | 72 | 10 | 48.3 | 0.036 * |
| M1 | 7 | 28 | 19 | 51.7 | |
| Age (n, %) | |||||
| ≤65 | 17 | 68 | 16 | 55.2 | 0.494 |
| >65 | 8 | 32 | 13 | 44.8 | |
| AFP (n, %) | |||||
| <400 ng/mL | 20 | 80 | 10 | 34.5 | 0.379 |
| ≥400 ng/mL | 5 | 20 | 19 | 65.5 | |
| SAE1 (n, %) | |||||
| <350 | 25 | 100 | 0 | 0 | <0.001 * |
| ≥350 | 0 | 0 | 29 | 100 | |
| Survival Status (n, %) | |||||
| Survived | 19 | 76 | 12 | 41.4 | 0.014 * |
| Expired | 2 | 8 | 11 | 37.9 | |
| Lost to follow-up | 4 | 16 | 6 | 20.7 | |
| Clinicopathological Variables | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| Gender Male vs. Female | 2.050 | 0.262–16.017 | 0.4937 | 0.575 | 0.063–5.278 | 0.6244 |
| Age, years ≤65 vs. >65 | 1.008 | 0.960–1.057 | 0.7532 | 0.968 | 0.922–1.017 | 0.1944 |
| AFP, ng/mL <400 vs. ≥400 | 2.375 | 0.725–7.782 | 0.1533 | 1.053 | 0.290–3.821 | 0.9378 |
| Metastasis M0 vs. M1 | 10.258 | 2.206–47.701 | 0.0030 * | 11.500 | 2.014–65.667 | 0.0060 * |
| SAE1 Q-Score <350 vs. ≥350 | 1.026 | 1.002–1.051 | 0.0319 * | 1.025 | 1.000–1.049 | 0.0468 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ong, J.R.; Bamodu, O.A.; Khang, N.V.; Lin, Y.-K.; Yeh, C.-T.; Lee, W.-H.; Cherng, Y.-G. SUMO-Activating Enzyme Subunit 1 (SAE1) Is a Promising Diagnostic Cancer Metabolism Biomarker of Hepatocellular Carcinoma. Cells 2021, 10, 178. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10010178
Ong JR, Bamodu OA, Khang NV, Lin Y-K, Yeh C-T, Lee W-H, Cherng Y-G. SUMO-Activating Enzyme Subunit 1 (SAE1) Is a Promising Diagnostic Cancer Metabolism Biomarker of Hepatocellular Carcinoma. Cells. 2021; 10(1):178. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10010178
Chicago/Turabian StyleOng, Jiann Ruey, Oluwaseun Adebayo Bamodu, Nguyen Viet Khang, Yen-Kuang Lin, Chi-Tai Yeh, Wei-Hwa Lee, and Yih-Giun Cherng. 2021. "SUMO-Activating Enzyme Subunit 1 (SAE1) Is a Promising Diagnostic Cancer Metabolism Biomarker of Hepatocellular Carcinoma" Cells 10, no. 1: 178. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10010178