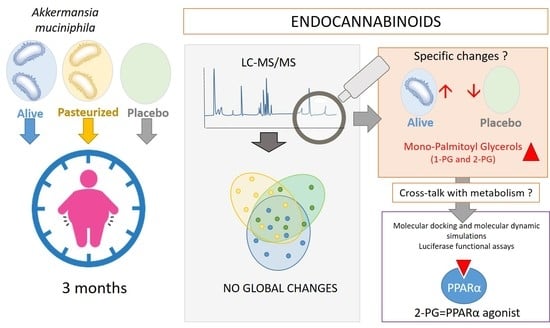
Beneficial Effects of Akkermansia muciniphila Are Not Associated with Major Changes in the Circulating Endocannabinoidome but Linked to Higher Mono-Palmitoyl-Glycerol Levels as New PPARα Agonists
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participant and Study Design
2.2. Biochemical Analysis
2.3. Computational Methods
2.4. Cell Culture, Transfection and Luciferase Assay
2.5. Statistical Analysis
3. Results
3.1. Preliminary Time-Series Analyses
3.2. Deeper Characterisation of the Effect of A. muciniphila on 1-PG and 2-PG Circulating Levels
3.3. Global eCBome Profiling Comparisons between Groups and Timing through Multivariate Analyses
3.4. In Silico Identification and In Vitro Validation of 2-PG as an Endogenous Activator of PPARα
3.5. In Silico Identification 1-PG as a Potential Endogenous Ligand of PPARα/γ
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dao, M.C.; Clement, K. Gut microbiota and obesity: Concepts relevant to clinical care. Eur. J. Intern. Med. 2018, 48, 18–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cani, P.D. Human gut microbiome: Hopes, threats and promises. Gut 2018, 67, 1716–1725. [Google Scholar] [CrossRef] [PubMed]
- Rastelli, M.; Knauf, C.; Cani, P.D. Gut Microbes and Health: A Focus on the Mechanisms Linking Microbes, Obesity, and Related Disorders. Obesity 2018, 26, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Everard, A. Talking microbes: When gut bacteria interact with diet and host organs. Mol. Nutr. Food Res. 2016, 60, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Belzer, C.; de Vos, W.M. Microbes inside--from diversity to function: The case of Akkermansia. ISME J. 2012, 6, 1449–1458. [Google Scholar] [CrossRef]
- Derrien, M.; Belzer, C.; de Vos, W.M. Akkermansia muciniphila and its role in regulating host functions. Microb. Pathog. 2017, 106, 171–181. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, N.; Tan, H.Y.; Li, S.; Zhang, C.; Feng, Y. Function of Akkermansia muciniphila in Obesity: Interactions with Lipid Metabolism, Immune Response and Gut Systems. Front. Microbiol. 2020, 11, 219. [Google Scholar] [CrossRef] [Green Version]
- Anhe, F.F.; Pilon, G.; Roy, D.; Desjardins, Y.; Levy, E.; Marette, A. Triggering Akkermansia with dietary polyphenols: A new weapon to combat the metabolic syndrome? Gut Microb. 2016, 7, 146–153. [Google Scholar] [CrossRef] [Green Version]
- Cani, P.D.; de Vos, W.M. Next-Generation Beneficial Microbes: The Case of Akkermansia muciniphila. Front. Microbiol. 2017, 8, 1765. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [Green Version]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef] [Green Version]
- Depommier, C.; Van Hul, M.; Everard, A.; Delzenne, N.M.; De Vos, W.M.; Cani, P.D. Pasteurized Akkermansia muciniphila increases whole-body energy expenditure and fecal energy excretion in diet-induced obese mice. Gut Microb. 2020, 11, 1231–1245. [Google Scholar] [CrossRef] [Green Version]
- Everard, A.; Plovier, H.; Rastelli, M.; Van Hul, M.; de Wouters d’Oplinter, A.; Geurts, L.; Druart, C.; Robine, S.; Delzenne, N.M.; Muccioli, G.G.; et al. Intestinal epithelial N-acylphosphatidylethanolamine phospholipase D links dietary fat to metabolic adaptations in obesity and steatosis. Nat. Commun. 2019, 10, 457. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.R.; Lee, J.C.; Lee, H.Y.; Kim, M.S.; Whon, T.W.; Lee, M.S.; Bae, J.W. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 2014, 63, 727–735. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Lin, S.; Vanhoutte, P.M.; Woo, C.W.; Xu, A. Akkermansia Muciniphila Protects Against Atherosclerosis by Preventing Metabolic Endotoxemia-Induced Inflammation in Apoe-/- Mice. Circulation 2016, 133, 2434–2446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Tong, X.; Sud, N.; Khound, R.; Song, Y.; Maldonado-Gomez, M.X.; Walter, J.; Su, Q. Low-Density Lipoprotein Receptor Signaling Mediates the Triglyceride-Lowering Action of Akkermansia muciniphila in Genetic-Induced Hyperlipidemia. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 1448–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanninen, A.; Toivonen, R.; Poysti, S.; Belzer, C.; Plovier, H.; Ouwerkerk, J.P.; Emani, R.; Cani, P.D.; De Vos, W.M. Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut 2017, 67, 1445–1453. [Google Scholar] [CrossRef] [Green Version]
- Grander, C.; Adolph, T.E.; Wieser, V.; Lowe, P.; Wrzosek, L.; Gyongyosi, B.; Ward, D.V.; Grabherr, F.; Gerner, R.R.; Pfister, A.; et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut 2018, 67, 891–901. [Google Scholar] [CrossRef]
- Wu, W.; Lv, L.; Shi, D.; Ye, J.; Fang, D.; Guo, F.; Li, Y.; He, X.; Li, L. Protective Effect of Akkermansia muciniphila against Immune-Mediated Liver Injury in a Mouse Model. Front. Microbiol. 2017, 8, 1804. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef]
- Di Marzo, V.; Izzo, A.A. Endocannabinoid overactivity and intestinal inflammation. Gut 2006, 55, 1373–1376. [Google Scholar] [CrossRef] [Green Version]
- McCoy, K.L. Interaction between Cannabinoid System and Toll-Like Receptors Controls Inflammation. Mediat. Inflamm. 2016, 2016, 5831315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellocchio, L.; Cervino, C.; Pasquali, R.; Pagotto, U. The endocannabinoid system and energy metabolism. J. Neuroendocrinol. 2008, 20, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Jager, G.; Witkamp, R.F. The endocannabinoid system and appetite: Relevance for food reward. Nutr. Res. Rev. 2014, 27, 172–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Marzo, V. New approaches and challenges to targeting the endocannabinoid system. Nat. Rev. Drug Discov. 2018, 17, 623–639. [Google Scholar] [CrossRef] [PubMed]
- Di Marzo, V.; Piscitelli, F. The Endocannabinoid System and its Modulation by Phytocannabinoids. Neurotherapeutics 2015, 12, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Di Marzo, V.; Wang, J.E. The Endocannabinoidome—The World of Endocannabinoids and Related Mediators; Academic Press: Cambridge, MA, USA, 2015; Volume 1, p. 208. [Google Scholar]
- Cani, P.D.; Plovier, H.; Van Hul, M.; Geurts, L.; Delzenne, N.M.; Druart, C.; Everard, A. Endocannabinoids—At the crossroads between the gut microbiota and host metabolism. Nat. Rev. Endocrinol. 2016, 12, 133–143. [Google Scholar] [CrossRef]
- Geurts, L.; Everard, A.; Van Hul, M.; Essaghir, A.; Duparc, T.; Matamoros, S.; Plovier, H.; Castel, J.; Denis, R.G.; Bergiers, M.; et al. Adipose tissue NAPE-PLD controls fat mass development by altering the browning process and gut microbiota. Nat. Commun. 2015, 6, 6495. [Google Scholar] [CrossRef] [Green Version]
- Di Marzo, V.; Côté, M.; Matias, I.; Lemieux, I.; Arsenault, B.J.; Cartier, A.; Piscitelli, F.; Petrosino, S.; Alméras, N.; Després, J.P. Changes in plasma endocannabinoid levels in viscerally obese men following a 1 year lifestyle modification programme and waist circumference reduction: Associations with changes in metabolic risk factors. Diabetologia 2009, 52, 213–217. [Google Scholar] [CrossRef] [Green Version]
- Di Marzo, V.; Verrijken, A.; Hakkarainen, A.; Petrosino, S.; Mertens, I.; Lundbom, N.; Piscitelli, F.; Westerbacka, J.; Soro-Paavonen, A.; Matias, I.; et al. Role of insulin as a negative regulator of plasma endocannabinoid levels in obese and nonobese subjects. Eur. J. Endocrinol. 2009, 161, 715–722. [Google Scholar] [CrossRef] [Green Version]
- Sipe, J.C.; Scott, T.M.; Murray, S.; Harismendy, O.; Simon, G.M.; Cravatt, B.F.; Waalen, J. Biomarkers of endocannabinoid system activation in severe obesity. PLoS ONE 2010, 5, e8792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castonguay-Paradis, S.; Lacroix, S.; Rochefort, G.; Parent, L.; Perron, J.; Martin, C.; Lamarche, B.; Raymond, F.; Flamand, N.; Di Marzo, V.; et al. Dietary fatty acid intake and gut microbiota determine circulating endocannabinoidome signaling beyond the effect of body fat. Sci. Rep. 2020, 10, 15975. [Google Scholar] [CrossRef] [PubMed]
- Osei-Hyiaman, D.; Liu, J.; Zhou, L.; Godlewski, G.; Harvey-White, J.; Jeong, W.I.; Bátkai, S.; Marsicano, G.; Lutz, B.; Buettner, C.; et al. Hepatic CB1 receptor is required for development of diet-induced steatosis, dyslipidemia, and insulin and leptin resistance in mice. J. Clin. Investig. 2008, 118, 3160–3169. [Google Scholar] [CrossRef] [PubMed]
- Osei-Hyiaman, D.; DePetrillo, M.; Pacher, P.; Liu, J.; Radaeva, S.; Bátkai, S.; Harvey-White, J.; Mackie, K.; Offertáler, L.; Wang, L.; et al. Endocannabinoid activation at hepatic CB1 receptors stimulates fatty acid synthesis and contributes to diet-induced obesity. J. Clin. Invest. 2005, 115, 1298–1305. [Google Scholar] [CrossRef] [Green Version]
- Silvestri, C.; Di Marzo, V. The endocannabinoid system in energy homeostasis and the etiopathology of metabolic disorders. Cell Metab. 2013, 17, 475–490. [Google Scholar] [CrossRef] [Green Version]
- Muccioli, G.G.; Naslain, D.; Backhed, F.; Reigstad, C.S.; Lambert, D.M.; Delzenne, N.M.; Cani, P.D. The endocannabinoid system links gut microbiota to adipogenesis. Mol. Syst. Biol. 2010, 6, 392. [Google Scholar] [CrossRef]
- Mehrpouya-Bahrami, P.; Chitrala, K.N.; Ganewatta, M.S.; Tang, C.; Murphy, E.A.; Enos, R.T.; Velazquez, K.T.; McCellan, J.; Nagarkatti, M.; Nagarkatti, P. Blockade of CB1 cannabinoid receptor alters gut microbiota and attenuates inflammation and diet-induced obesity. Sci. Rep. 2017, 7, 15645. [Google Scholar] [CrossRef]
- Van Gaal, L.F.; Rissanen, A.M.; Scheen, A.J.; Ziegler, O.; Rossner, S.; Group, R.I.-E.S. Effects of the cannabinoid-1 receptor blocker rimonabant on weight reduction and cardiovascular risk factors in overweight patients: 1-year experience from the RIO-Europe study. Lancet 2005, 365, 1389–1397. [Google Scholar] [CrossRef]
- Lacroix, S.; Pechereau, F.; Leblanc, N.; Boubertakh, B.; Houde, A.; Martin, C.; Flamand, N.; Silvestri, C.; Raymond, F.; Di Marzo, V.; et al. Rapid and Concomitant Gut Microbiota and Endocannabinoidome Response to Diet-Induced Obesity in Mice. mSystems 2019, 4. [Google Scholar] [CrossRef] [Green Version]
- Bian, X.; Wu, W.; Yang, L.; Lv, L.; Wang, Q.; Li, Y.; Ye, J.; Fang, D.; Wu, J.; Jiang, X.; et al. Administration of Akkermansia muciniphila Ameliorates Dextran Sulfate Sodium-Induced Ulcerative Colitis in Mice. Front. Microbiol. 2019, 10, 2259. [Google Scholar] [CrossRef] [Green Version]
- Tutunchi, H.; Saghafi-Asl, M.; Ostadrahimi, A. A systematic review of the effects of oleoylethanolamide, a high-affinity endogenous ligand of PPAR-α, on the management and prevention of obesity. Clin. Exp. Pharm. Physiol. 2020, 47, 543–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, L.; Shen, W.-J.; Bittner, S.; Kraemer, F.B.; Azhar, S. PPARs: Regulators of metabolism and as therapeutic targets in cardiovascular disease. Part I: PPAR-α. Future Cardiol. 2017, 13, 259–278. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, C.; Krey, G.; Keller, H.; Givel, F.; Helftenbein, G.; Wahli, W. Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell 1992, 68, 879–887. [Google Scholar] [CrossRef]
- Ribet, C.; Montastier, E.; Valle, C.; Bezaire, V.R.; Mazzucotelli, A.; Mairal, A.; Viguerie, N.; Langin, D. Peroxisome Proliferator-Activated Receptor-α Control of Lipid and Glucose Metabolism in Human White Adipocytes. Endocrinology 2010, 151, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Lamichane, S.; Dahal Lamichane, B.; Kwon, S.-M. Pivotal Roles of Peroxisome Proliferator-Activated Receptors (PPARs) and Their Signal Cascade for Cellular and Whole-Body Energy Homeostasis. Int. J. Mol. Sci. 2018, 19, 949. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Su, Y.; Silva, F.J.; Weller, A.H.; Sostre-Colón, J.; Titchenell, P.M.; Steger, D.J.; Seale, P.; Soccio, R.E. Shared PPARα/γ Target Genes Regulate Brown Adipocyte Thermogenic Function. Cell Rep. 2020, 30, 3079–3091. [Google Scholar] [CrossRef] [Green Version]
- Lo Verme, J.; Fu, J.; Astarita, G.; La Rana, G.; Russo, R.; Calignano, A.; Piomelli, D. The nuclear receptor peroxisome proliferator-activated receptor-alpha mediates the anti-inflammatory actions of palmitoylethanolamide. Mol. Pharmacol. 2005, 67, 15–19. [Google Scholar] [CrossRef]
- Karwad, M.A.; Macpherson, T.; Wang, B.; Theophilidou, E.; Sarmad, S.; Barrett, D.A.; Larvin, M.; Wright, K.L.; Lund, J.N.; O’Sullivan, S.E. Oleoylethanolamine and palmitoylethanolamine modulate intestinal permeability in vitro via TRPV1 and PPARalpha. FASEB J. 2017, 31, 469–481. [Google Scholar] [CrossRef] [Green Version]
- Bouaboula, M.; Hilairet, S.; Marchand, J.; Fajas, L.; Le Fur, G.; Casellas, P. Anandamide induced PPARgamma transcriptional activation and 3T3-L1 preadipocyte differentiation. Eur. J. Pharmacol. 2005, 517, 174–181. [Google Scholar] [CrossRef]
- Tontonoz, P.; Spiegelman, B.M. Fat and beyond: The diverse biology of PPARgamma. Ann. Rev. Biochem. 2008, 77, 289–312. [Google Scholar] [CrossRef]
- Werner, M.; Tonjes, A.; Stumvoll, M.; Thiery, J.; Kratzsch, J. Assay-dependent variability of serum insulin levels during oral glucose tolerance test: Influence on reference intervals for insulin and on cut-off values for insulin sensitivity indices. Clin. Chem. Lab. Med. 2008, 46, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Borza, D.; Karmali, R.; Andre, S.; Valsamis, J.; Cytryn, E.; Hermans, M.P.; Beyer, I. Influence of assay-dependent variability of serum insulin levels on insulin sensitivity indices. Clin. Chem. Lab. Med. 2008, 46, 1655–1656. [Google Scholar] [CrossRef] [PubMed]
- Turcotte, C.; Archambault, A.S.; Dumais, É.; Martin, C.; Blanchet, M.R.; Bissonnette, E.; Ohashi, N.; Yamamoto, K.; Itoh, T.; Laviolette, M.; et al. Endocannabinoid hydrolysis inhibition unmasks that unsaturated fatty acids induce a robust biosynthesis of 2-arachidonoyl-glycerol and its congeners in human myeloid leukocytes. FASEB J. 2020, 34, 4253–4265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acton, A.; Bank, M.; Bréfort, J.; Cruz, M.; Curtis, D.; Hassinen, T.; Heikkilä, V.; Hutchison, G.; Huuskonen, J.; Jensen, J.; et al. Ghemical; Department of Chemistry, University of Kuopio: Kuopio, Finland, 2006. [Google Scholar]
- Clark, M.; Cramer Iii, R.D.; Van Opdenbosch, N. Validation of the general purpose tripos 5.2 force field. J. Comput. Chem. 1989, 10, 982–1012. [Google Scholar] [CrossRef]
- Schmidt, M.W.; Baldridge, K.K.; Boatz, J.A.; Elbert, S.T.; Gordon, M.S.; Jensen, J.H.; Koseki, S.; Matsunaga, N.; Nguyen, K.A.; Su, S.; et al. General atomic and molecular electronic structure system. J. Comput. Chem. 1993, 14, 1347–1363. [Google Scholar] [CrossRef]
- Fox, T.; Kollman, P.A. Application of the RESP Methodology in the Parametrization of Organic Solvents. J. Phys. Chem. B 1998, 102, 8070–8079. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Iannotti, F.A.; De Maio, F.; Panza, E.; Appendino, G.; Taglialatela-Scafati, O.; De Petrocellis, L.; Amodeo, P.; Vitale, R.M. Identification and Characterization of Cannabimovone, a Cannabinoid from Cannabis sativa, as a Novel PPARgamma Agonist via a Combined Computational and Functional Study. Molecules 2020, 25, 1119. [Google Scholar] [CrossRef] [Green Version]
- Case, D.A.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.E.; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Homeyer, N.; et al. AMBER 2016; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- D’Aniello, E.; Fellous, T.; Iannotti, F.A.; Gentile, A.; Allarà, M.; Balestrieri, F.; Gray, R.; Amodeo, P.; Vitale, R.M.; Di Marzo, V. Identification and characterization of phytocannabinoids as novel dual PPARα/γ agonists by a computational and in vitro experimental approach. Biochim. Biophys. Acta 2019, 1863, 586–597. [Google Scholar] [CrossRef]
- Canuto, R.; Camey, S.; Gigante, D.; Menezes, A.; Olinto, M.T. Focused Principal Component Analysis: A graphical method for exploring dietary patterns. Cad. Saude Publ. 2010, 26, 2149–2156. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Szymańska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veilleux, A.; Di Marzo, V.; Silvestri, C. The Expanded Endocannabinoid System/Endocannabinoidome as a Potential Target for Treating Diabetes Mellitus. Curr. Diab. Rep. 2019, 19, 117. [Google Scholar] [CrossRef] [PubMed]
- Di Marzo, V.; Silvestri, C. Lifestyle and Metabolic Syndrome: Contribution of the Endocannabinoidome. Nutrients 2019, 11, 1956. [Google Scholar] [CrossRef] [Green Version]
- Depommier, C.; Flamand, N.; Pelicaen, R.; Maiter, D.; Thissen, J.P.; Loumaye, A.; Hermans, M.P.; Everard, A.; Delzenne, N.; Di Marzo, V.; et al. Linking the Endocannabinoidome with Specific Metabolic Parameters in an Overweight and Insulin-Resistant Population: From Multivariate Exploratory Analysis to Univariate Analysis and Construction of Predictive Models. Cells 2021, 10, 71. [Google Scholar] [CrossRef]
- Poursharifi, P.; Madiraju, S.R.M.; Prentki, M. Monoacylglycerol signalling and ABHD6 in health and disease. Diabetes Obes. Metab. 2017, 19, 76–89. [Google Scholar] [CrossRef] [Green Version]
- Pistis, M.; O’Sullivan, S.E. The Role of Nuclear Hormone Receptors in Cannabinoid Function. Adv. Pharmacol. 2017, 80, 291–328. [Google Scholar] [CrossRef]
- Pagano, C.; Rossato, M.; Vettor, R. Endocannabinoids, Adipose Tissue and Lipid Metabolism. J. Neuroendocrinol. 2008, 20, 124–129. [Google Scholar] [CrossRef]
- Fu, J.; Gaetani, S.; Oveisi, F.; Lo Verme, J.; Serrano, A.; Rodriguez De Fonseca, F.; Rosengarth, A.; Luecke, H.; Di Giacomo, B.; Tarzia, G.; et al. Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-alpha. Nature 2003, 425, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Oveisi, F.; Gaetani, S.; Lin, E.; Piomelli, D. Oleoylethanolamide, an endogenous PPAR-alpha agonist, lowers body weight and hyperlipidemia in obese rats. Neuropharmacology 2005, 48, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez de Fonseca, F.; Navarro, M.; Gomez, R.; Escuredo, L.; Nava, F.; Fu, J.; Murillo-Rodriguez, E.; Giuffrida, A.; LoVerme, J.; Gaetani, S.; et al. An anorexic lipid mediator regulated by feeding. Nature 2001, 414, 209–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzmán, M.; Lo Verme, J.; Fu, J.; Oveisi, F.; Blázquez, C.; Piomelli, D. Oleoylethanolamide stimulates lipolysis by activating the nuclear receptor peroxisome proliferator-activated receptor alpha (PPAR-alpha). J. Biol. Chem. 2004, 279, 27849–27854. [Google Scholar] [CrossRef] [Green Version]
- Zygmunt, P.M.; Ermund, A.; Movahed, P.; Andersson, D.A.; Simonsen, C.; Jönsson, B.A.G.; Blomgren, A.; Birnir, B.; Bevan, S.; Eschalier, A.; et al. Monoacylglycerols Activate TRPV1—A Link between Phospholipase C and TRPV1. PLoS ONE 2013, 8, e81618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierre, J.F.; Martinez, K.B.; Ye, H.; Nadimpalli, A.; Morton, T.C.; Yang, J.; Wang, Q.; Patno, N.; Chang, E.B.; Yin, D.P. Activation of bile acid signaling improves metabolic phenotypes in high-fat diet-induced obese mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 311, 286–304. [Google Scholar] [CrossRef] [Green Version]
- Anhe, F.F.; Varin, T.V.; Le Barz, M.; Pilon, G.; Dudonne, S.; Trottier, J.; St-Pierre, P.; Harris, C.S.; Lucas, M.; Lemire, M.; et al. Arctic berry extracts target the gut-liver axis to alleviate metabolic endotoxaemia, insulin resistance and hepatic steatosis in diet-induced obese mice. Diabetologia 2018, 61, 919–931. [Google Scholar] [CrossRef] [Green Version]
- Ashrafian, F.; Shahriary, A.; Behrouzi, A.; Moradi, H.R.; Keshavarz Azizi Raftar, S.; Lari, A.; Hadifar, S.; Yaghoubfar, R.; Ahmadi Badi, S.; Khatami, S.; et al. Akkermansia muciniphila-Derived Extracellular Vesicles as a Mucosal Delivery Vector for Amelioration of Obesity in Mice. Front. Microbiol. 2019, 10, 2155. [Google Scholar] [CrossRef]






| Endocannabinoids and Related Mediators | |
|---|---|
| 1-PG | 2-palmitoyl-glycerol |
| 2-PG | 1-palmitoyl-glycerol |
| 1-PG & 2-PG | 1-palmitoyl-glycerol + 2-palmitoyl-glycerol |
| 2-OG | 1(3)- and 2-oleoylglycerol |
| 2-LG | 1(3)- and 2-linoleoyl-glycerol |
| 2-AG | 1(3)- and 2-arachidonoyl-glycerol |
| 2-DHG | 1(3)- and 2-docosahexaenoyl-glycerol |
| 2-EPG | 1(3)- and 2- eicosapentaenoyl-glycerol |
| 2-DPG | 1(3)- and 2-docospentaenoyl-glycerol(n − 3) |
| PEA | N-palmitoyl-ethanolamine |
| SEA | N-stearoyl-ethanolamine |
| OEA | N-oleoyl-ethanolamine |
| LEA | N-linoleoyl-ethanolamine |
| AEA | N-arachidonoyl-ethanolamine |
| EPEA | N-eicosapentaenoyl-ethanolamine |
| DPEA | N-docosapentaenoyl-ethanolamine(n − 3) |
| DHEA | N-docosahexanoyl-ethanolamine |
| AA | arachidonic acid |
| EPA | eicosapentaenoic acid |
| DPA | docosapentaenoic acid (n − 3) |
| DHA | docosahexaenoic acid |
| Measure | 1 Comps | 2 Comps | 3 Comps | 4 Comps | 5 Comps |
|---|---|---|---|---|---|
| Accuracy | 0.374 | 0.313 | 0.344 | 0.344 | 0.313 |
| R2 a | 0.14165 | 0.339 | 0.382 | 0.410 | 0.427 |
| Q2 b | −0.1494 | −0.852 | −1.015 | −1.309 | −1.985 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Depommier, C.; Vitale, R.M.; Iannotti, F.A.; Silvestri, C.; Flamand, N.; Druart, C.; Everard, A.; Pelicaen, R.; Maiter, D.; Thissen, J.-P.; et al. Beneficial Effects of Akkermansia muciniphila Are Not Associated with Major Changes in the Circulating Endocannabinoidome but Linked to Higher Mono-Palmitoyl-Glycerol Levels as New PPARα Agonists. Cells 2021, 10, 185. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10010185
Depommier C, Vitale RM, Iannotti FA, Silvestri C, Flamand N, Druart C, Everard A, Pelicaen R, Maiter D, Thissen J-P, et al. Beneficial Effects of Akkermansia muciniphila Are Not Associated with Major Changes in the Circulating Endocannabinoidome but Linked to Higher Mono-Palmitoyl-Glycerol Levels as New PPARα Agonists. Cells. 2021; 10(1):185. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10010185
Chicago/Turabian StyleDepommier, Clara, Rosa Maria Vitale, Fabio Arturo Iannotti, Cristoforo Silvestri, Nicolas Flamand, Céline Druart, Amandine Everard, Rudy Pelicaen, Dominique Maiter, Jean-Paul Thissen, and et al. 2021. "Beneficial Effects of Akkermansia muciniphila Are Not Associated with Major Changes in the Circulating Endocannabinoidome but Linked to Higher Mono-Palmitoyl-Glycerol Levels as New PPARα Agonists" Cells 10, no. 1: 185. https://0-doi-org.brum.beds.ac.uk/10.3390/cells10010185