SETDB1 Regulates Porcine Spermatogonial Adhesion and Proliferation through Modulating MMP3/10 Transcription
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Animals
2.3. Immunofluorescence Analysis
2.4. Cell Culture
2.5. SETDB1-siRNA Transfection
2.6. qRT-PCR Analysis
2.7. Western Blot Analysis
2.8. EdU Staining Assay
2.9. Cell Cycle Analysis
2.10. Cell-Adhesion Assay
2.11. StatisticalAanalysis
3. Results
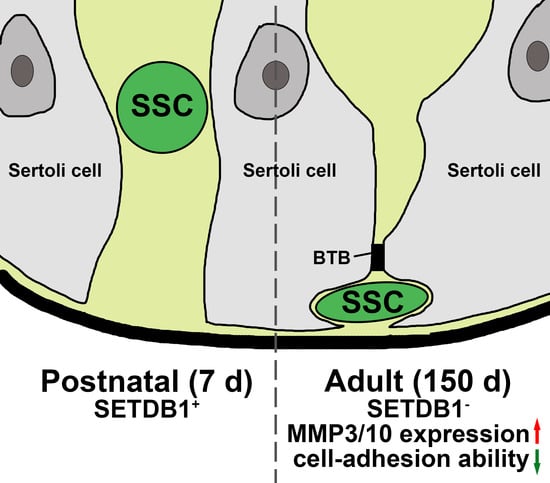
3.1. Characterization of SETDB1 during Porcine Spermatogenesis
3.2. Knockdown (KD) of SETDB1 via siRNAs in Porcine SSCs
3.3. SETDB1-KD Impaired Cell Proliferation and Cell Cycle Transition
3.4. SETDB1-KD Affected Cell Adhesion and Cell Spread
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kanatsu-Shinohara, M.; Ogonuki, N.; Iwano, T.; Lee, J.; Kazuki, Y.; Inoue, K.; Miki, H.; Takehashi, M.; Toyokuni, S.; Shinkai, Y.; et al. Genetic and epigenetic properties of mouse male germline stem cells during long-term culture. Development 2005, 132, 4155–4163. [Google Scholar] [CrossRef] [Green Version]
- Kubota, H.; Brinster, R.L. Spermatogonial stem cells. Biol. Reprod. 2018, 99, 52–74. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.-Y.; Willis, W.D.; Eddy, E.M. Targeting the Gdnf Gene in peritubular myoid cells disrupts undifferentiated spermatogonial cell development. Proc. Natl. Acad. Sci. USA 2016, 113, 1829–1834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Rooij, D.G. The spermatogonial stem cell niche. Microsc. Res. Tech. 2009, 72, 580–585. [Google Scholar] [CrossRef]
- Oatley, M.J.; Racicot, K.E.; Oatley, J.M. Sertoli cells dictate spermatogonial stem cell niches in the mouse testis. Biol. Reprod. 2011, 84, 639–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, Y.; Liu, S.; Guan, Y.; Pan, H.; Guan, X.; Qiu, Z.; Li, L.; Gao, N.; Zhao, Y.; Li, X.; et al. Lgr4-mediated Wnt/β-catenin signaling in peritubular myoid cells is essential for spermatogenesis. Development 2013, 140, 1751–1761. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.; Lindahl, M.; Hyvonen, M.E.; Parvinen, M.; de Rooij, D.G.; Hess, M.W.; Raatikainen-Ahokas, A.; Sainio, K.; Rauvala, H.; Lakso, M.; et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 2000, 287, 1489–1493. [Google Scholar] [CrossRef]
- Takase, H.M.; Nusse, R. Paracrine Wnt/β-catenin signaling mediates proliferation of undifferentiated spermatogonia in the adult mouse testis. Proc. Natl. Acad. Sci. USA 2016, 113, E1489–E1497. [Google Scholar] [CrossRef] [Green Version]
- Maekawa, M.; Kamimura, K.; Nagano, T. Peritubular myoid cells in the testis: Their structure and function. Arch. Histol. Cytol. 1996, 59, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kokkinaki, M.; Lee, T.-L.; He, Z.; Jiang, J.; Golestaneh, N.; Hofmann, M.-C.; Chan, W.-Y.; Dym, M. The molecular signature of spermatogonial stem/progenitor cells in the 6-day-old mouse testis. Biol. Reprod. 2009, 80, 707–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanatsu-Shinohara, M.; Takehashi, M.; Takashima, S.; Lee, J.; Morimoto, H.; Chuma, S.; Raducanu, A.; Nakatsuji, N.; Fässler, R.; Shinohara, T. Homing of Mouse Spermatogonial Stem Cells to Germline Niche Depends on beta1-Integrin. Cell Stem Cell 2008, 3, 533–542. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Jiang, J.; Kokkinaki, M.; Golestaneh, N.; Hofmann, M.-C.; Dym, M. Gdnf upregulates c-Fos transcription via the Ras/Erk1/2 pathway to promote mouse spermatogonial stem cell proliferation. Stem Cells 2008, 26, 266–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oatley, J.M.; Avarbock, M.R.; Brinster, R.L. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J. Biol. Chem. 2007, 282, 25842–25851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, M.; Braun, R.E. Cyclical expression of GDNF is required for spermatogonial stem cell homeostasis. Development 2018, 145, dev.151555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakiuchi, K.; Taniguchi, K.; Kubota, H. Conserved and non-conserved characteristics of porcine glial cell line-derived neurotrophic factor expressed in the testis. Sci. Rep. 2018, 8, 7656. [Google Scholar] [CrossRef]
- McSwiggin, H.M.; O’Doherty, A.M. Epigenetic reprogramming during spermatogenesis and male factor infertility. Reproduction 2018, 156, R9–R21. [Google Scholar] [CrossRef] [PubMed]
- Hammoud, S.S.; Low, D.H.P.; Yi, C.; Carrell, D.T.; Guccione, E.; Cairns, B.R. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 2014, 15, 239–253. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, K.; Doi, S.; Nakashima, A.; Irifuku, T.; Yamada, K.; Kokoroishi, K.; Ueno, T.; Doi, T.; Hida, E.; Arihiro, K.; et al. Inhibition of SET Domain-Containing Lysine Methyltransferase 7/9 Ameliorates Renal Fibrosis. J. Am. Soc. Nephrol. 2016, 27, 203–215. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Grow, E.J.; Mlcochova, H.; Maher, G.J.; Lindskog, C.; Nie, X.; Guo, Y.; Takei, Y.; Yun, J.; Cai, L.; et al. The adult human testis transcriptional cell atlas. Cell Res. 2018, 28, 1141–1157. [Google Scholar] [CrossRef] [PubMed]
- Maezawa, S.; Yukawa, M.; Alavattam, K.G.; Barski, A.; Namekawa, S.H. Dynamic reorganization of open chromatin underlies diverse transcriptomes during spermatogenesis. Nucleic Acids Res. 2018, 46, 593–608. [Google Scholar] [CrossRef] [Green Version]
- Tomizawa, S.-I.; Kobayashi, Y.; Shirakawa, T.; Watanabe, K.; Mizoguchi, K.; Hoshi, I.; Nakajima, K.; Nakabayashi, J.; Singh, S.; Dahl, A.; et al. Kmt2b conveys monovalent and bivalent H3K4me3 in mouse spermatogonial stem cells at germline and embryonic promoters. Development 2018, 145, dev.169102. [Google Scholar] [CrossRef] [Green Version]
- Lambrot, R.; Lafleur, C.; Kimmins, S. The histone demethylase KDM1A is essential for the maintenance and differentiation of spermatogonial stem cells and progenitors. FASEB J. 2015, 29, 4402–4416. [Google Scholar] [CrossRef]
- Kato, M.; Takemoto, K.; Shinkai, Y. A somatic role for the histone methyltransferase Setdb1 in endogenous retrovirus silencing. Nat. Commun. 2018, 9, 1683. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Brind’Amour, J.; Karimi, M.M.; Shirane, K.; Bogutz, A.; Lefebvre, L.; Sasaki, H.; Shinkai, Y.; Lorincz, M.C. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Gene Dev. 2014, 28, 2041–2055. [Google Scholar] [CrossRef] [Green Version]
- Hirota, T.; Blakeley, P.; Sangrithi, M.N.; Mahadevaiah, S.K.; Encheva, V.; Snijders, A.P.; ElInati, E.; Ojarikre, O.A.; de Rooij, D.G.; Niakan, K.K.; et al. SETDB1 Links the Meiotic DNA Damage Response to Sex Chromosome Silencing in Mice. Dev. Cell 2018, 47, 645–659. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Ye, F.; Li, Y.-Y.; Zhan, Y.-Z.; Liu, Y.; Yan, H.-M.; Fang, Y.; Xie, Y.-W.; Zhang, F.-J.; Chen, L.-H.; et al. Histone methyltransferase SETDB1 promotes colorectal cancer proliferation through the STAT1-CCND1/CDK6 axis. Carcinogenesis 2020, 41, 678–688. [Google Scholar] [CrossRef]
- Huang, J.; Huang, W.; Liu, M.; Zhu, J.; Jiang, D.; Xiong, Y.; Zhen, Y.; Yang, D.; Chen, Z.; Peng, L.; et al. Enhanced expression of SETDB1 possesses prognostic value and promotes cell proliferation, migration and invasion in nasopharyngeal carcinoma. Oncol. Rep. 2018, 40, 1017–1025. [Google Scholar] [CrossRef]
- Spyropoulou, A.; Gargalionis, A.; Dalagiorgou, G.; Adamopoulos, C.; Papavassiliou, K.A.; Lea, R.W.; Piperi, C.; Papavassiliou, A.G. Role of histone lysine methyltransferases SUV39H1 and SETDB1 in gliomagenesis: Modulation of cell proliferation, migration, and colony formation. Neuromolecular. Med. 2014, 16, 70–82. [Google Scholar] [CrossRef]
- An, J.; Zhang, X.; Qin, J.; Wan, Y.; Hu, Y.; Liu, T.; Li, J.; Dong, W.; Du, E.; Pan, C.; et al. The histone methyltransferase ESET is required for the survival of spermatogonial stem/progenitor cells in mice. Cell Death. Dis. 2014, 5, e1196. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Chen, X.; Li, T.; Li, X.; Lyu, Y.; Fan, X.; Zhang, P.; Zeng, W. Histone methyltransferase SETDB1 maintains survival of mouse spermatogonial stem/progenitor cells via PTEN/AKT/FOXO1 pathway. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 1094–1102. [Google Scholar] [CrossRef]
- Zheng, Y.; Feng, T.; Zhang, P.; Lei, P.; Li, F.; Zeng, W. Establishment of cell lines with porcine spermatogonial stem cell properties. J. Anim. Sci. Biotechnol. 2020, 11, 33. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Chen, X.; Zhang, P.; Li, F.; Zhang, L.; Li, X.; Huang, T.; Zheng, Y.; Yu, T.; Zhang, T.; et al. Transcriptome-wide Dynamics of m6A mRNA Methylation During Porcine Spermatogenesis. Genomics Proteomics Bioinf. 2021. [Google Scholar] [CrossRef]
- Luo, J.; Megee, S.; Dobrinski, I. Asymmetric distribution of UCH-L1 in spermatogonia is associated with maintenance and differentiation of spermatogonial stem cells. J. Cell Physiol. 2009, 220, 460–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almunia, J.; Nakamura, K.; Murakami, M.; Takashima, S.; Takasu, M. Characterization of domestic pig spermatogenesis using spermatogonial stem cell markers in the early months of life. Theriogenology 2018, 107, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Megee, S.; Rathi, R.; Dobrinski, I. Protein gene product 9.5 is a spermatogonia-specific marker in the pig testis: Application to enrichment and culture of porcine spermatogonia. Mol. Reprod. Dev. 2006, 73, 1531–1540. [Google Scholar] [CrossRef]
- Yuan, L.; Liu, J.G.; Zhao, J.; Brundell, E.; Daneholt, B.; Höög, C. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell 2000, 5, 73–83. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, P.; Li, T.; Chen, X.; Zhu, Z.; Lyu, Y.; Li, X.; Tian, X.; Zeng, W. SETDB1 plays an essential role in maintenance of gonocyte survival in pigs. Reproduction 2017, 154, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Shang, W.; Wang, Y.; Liang, X.; Li, T.; Shao, W.; Liu, F.; Cui, X.; Wang, Y.; Lv, L.; Chai, L.; et al. SETDB1 promotes gastric carcinogenesis and metastasis via upregulation of CCND1 and MMP9 expression. J. Pathol. 2021, 253, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Ceol, C.J.; Houvras, Y.; Jane-Valbuena, J.; Bilodeau, S.; Orlando, D.A.; Battisti, V.; Fritsch, L.; Lin, W.M.; Hollmann, T.J.; Ferré, F.; et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 2011, 471, 513–517. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.-M.; Wei, L.; Law, C.-T.; Ho, D.W.-H.; Tsang, F.H.-C.; Au, S.L.-K.; Sze, K.M.-F.; Lee, J.M.-F.; Wong, C.C.-L.; Ng, I.O.-L. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology 2016, 63, 474–487. [Google Scholar] [CrossRef] [Green Version]
- Bates, M.; Furlong, F.; Gallagher, M.F.; Spillane, C.D.; McCann, A.; O’Toole, S.; O’Leary, J.J. Too MAD or not MAD enough: The duplicitous role of the spindle assembly checkpoint protein MAD2 in cancer. Cancer Lett. 2020, 469, 11–21. [Google Scholar] [CrossRef]
- Sun, S.-C.; Kim, N.-H. Spindle assembly checkpoint and its regulators in meiosis. Hum. Reprod. Update 2012, 18, 60–72. [Google Scholar] [CrossRef] [Green Version]
- Siu, M.K.Y.; Cheng, C.Y. Extracellular matrix and its role in spermatogenesis. Adv. Exp. Med. Biol. 2008, 636, 74–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cappelli, C.; Sepulveda, H.; Rivas, S.; Pola, V.; Urzúa, U.; Donoso, G.; Sagredo, E.; Carrero, D.; Casanova-Ortiz, E.; Sagredo, A.; et al. Ski Is Required for Tri-Methylation of H3K9 in Major Satellite and for Repression of Pericentromeric Genes: Mmp3, Mmp10 and Mmp13, in Mouse Fibroblasts. J. Mol. Biol. 2020, 432, 3222–3238. [Google Scholar] [CrossRef]
- Chen, K.; Zhang, F.; Ding, J.; Liang, Y.; Zhan, Z.; Zhan, Y.; Chen, L.-H.; Ding, Y. Histone Methyltransferase SETDB1 Promotes the Progression of Colorectal Cancer by Inhibiting the Expression of TP53. J. Cancer 2017, 8, 3318–3330. [Google Scholar] [CrossRef] [Green Version]
- Na, H.-H.; Noh, H.-J.; Cheong, H.-M.; Kang, Y.; Kim, K.-C. SETDB1 mediated FosB expression increases the cell proliferation rate during anticancer drug therapy. BMB Rep. 2016, 49, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Jurkowska, R.Z.; Qin, S.; Kungulovski, G.; Tempel, W.; Liu, Y.; Bashtrykov, P.; Stiefelmaier, J.; Jurkowski, T.P.; Kudithipudi, S.; Weirich, S.; et al. H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1. Nat. Commun. 2017, 8, 2057. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Xia, L.; Wu, D.Y.; Wang, H.; Chansky, H.A.; Schubach, W.H.; Hickstein, D.D.; Zhang, Y. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene 2002, 21, 148–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Long, J.; Gao, Y.; Zhang, W.; Han, F.; Xu, C.; Sun, L.; Yang, S.-C.; Lan, J.; Hou, Z.; et al. SETDB1-mediated methylation of Akt promotes itsK63-linked ubiquitination and activation leading totumorigenesis. Nat. Cell Biol. 2019, 21, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Brinster, R.L.; Zimmermann, J.W. Spermatogenesis following male germ-cell transplantation. Proc. Natl. Acad. Sci. USA 1994, 91, 11298–11302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tokuda, M.; Kadokawa, Y.; Kurahashi, H.; Marunouchi, T. CDH1 is a specific marker for undifferentiated spermatogonia in mouse testes. Biol. Reprod. 2007, 76, 130–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bendel-Stenzel, M.R.; Gomperts, M.; Anderson, R.; Heasman, J.; Wylie, C. The role of cadherins during primordial germ cell migration and early gonad formation in the mouse. Mech. Dev. 2000, 91, 143–152. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.; Liu, Z.; Guo, M.; Zeng, W.; Zheng, Y. SETDB1 Regulates Porcine Spermatogonial Adhesion and Proliferation through Modulating MMP3/10 Transcription. Cells 2022, 11, 370. https://0-doi-org.brum.beds.ac.uk/10.3390/cells11030370
Liu R, Liu Z, Guo M, Zeng W, Zheng Y. SETDB1 Regulates Porcine Spermatogonial Adhesion and Proliferation through Modulating MMP3/10 Transcription. Cells. 2022; 11(3):370. https://0-doi-org.brum.beds.ac.uk/10.3390/cells11030370
Chicago/Turabian StyleLiu, Ruifang, Zidong Liu, Ming Guo, Wenxian Zeng, and Yi Zheng. 2022. "SETDB1 Regulates Porcine Spermatogonial Adhesion and Proliferation through Modulating MMP3/10 Transcription" Cells 11, no. 3: 370. https://0-doi-org.brum.beds.ac.uk/10.3390/cells11030370