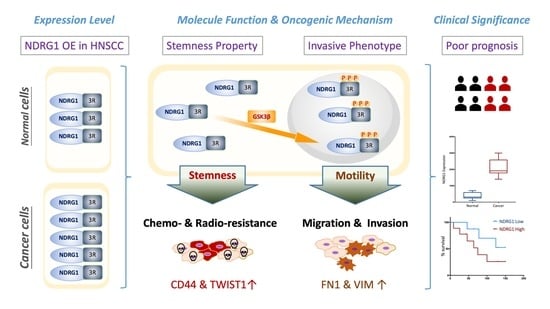
Multifaceted and Intricate Oncogenic Mechanisms of NDRG1 in Head and Neck Cancer Depend on Its C-Terminal 3R-Motif
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cellular Transfection
2.2. Construction of Full-Length and Truncated NDRG1 Expression Plasmids
2.3. Transcriptomic and Bioinformatic Analyses in a Global Survey of Molecular Functions
2.4. Assessment of Cancer Stemness by Spheroid Cell Formation
2.5. Determination of Radiosensitivity and Chemosensitivity
2.6. Analyses of Cell Invasion and Migration
2.7. RNA Extraction and RT-qPCR Analysis
2.8. Protein Extraction, Subcellular Fractionation, and Western Blot Analysis
2.9. Immunofluorescence Analysis and Confocal Microscopic Examination
2.10. Immunoprecipitation, Kinase Assay, and In Vitro Assessment of Phosphorylation
2.11. Clinical Tissue Expression and Prognostic Evaluation of NDRG1 in Patients with HNSC
3. Results
3.1. Transcriptomic Analysis Reveals NDRG1 Participation in Diverse Oncogenic Functions
3.2. NDRG1 Contributes to Cancer Stemness and Leads to Chemo-Radioresistance
3.3. NDRG1 Facilitates Cell Motility via Modulation of Extracellular Matrix (ECM)
3.4. 3R-Motif of NDRG1 Has Minimal Effect on Cancer Stemness
3.5. 3R-Motif of NDRG1 Is Critical for Cell Motility
3.6. 3R-Motif of NDRG1 Is Required for Nuclear Translocation and Phosphorylation
3.7. Phosphorylation of the 3R-Motif by GSK3b Is Critical for Cell Motility in HNC Cells
3.8. NDRG1 Is Overexpressed in Tumors and Is Associated with Poor Prognosis in Patients with HNC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Meltzer, C.; Nguyen, N.T.; Zhang, J.; Aguilar, J.; Blatchins, M.A.; Quesenberry, C.P., Jr.; Wang, Y.; Sakoda, L.C. Survival associated with consolidated multidisciplinary care in head and neck cancer: A retrospective cohort study. Otolaryngol. Head Neck Surg. 2021, 9, 1945998211057852. [Google Scholar] [CrossRef]
- Chang, J.T.; Wang, H.M.; Chang, K.W.; Chen, W.H.; Wen, M.C.; Hsu, Y.M.; Yung, B.Y.; Chen, I.H.; Liao, C.T.; Hsieh, L.L.; et al. Identification of differentially expressed genes in oral squamous cell carcinoma (OSCC): Overexpression of NPM, CDK1 and NDRG1 and underexpression of CHES1. Int. J. Cancer 2005, 114, 942–949. [Google Scholar] [CrossRef]
- Lorini, L.; Ardighieri, L.; Bozzola, A.; Romani, C.; Bignotti, E.; Buglione, M.; Guerini, A.; Lombardi, D.; Deganello, A.; Tomasoni, M.; et al. Prognosis and management of recurrent and/or metastatic head and neck adenoid cystic carcinoma. Oral Oncol. 2021, 115, 105213. [Google Scholar] [CrossRef]
- Tang, E.; Nguyen, T.V.; Clatot, F.; Rambeau, A.; Johnson, A.; Sun, X.S.; Tao, Y.; Thariat, J. Radiation therapy on primary tumour of synchronous metastatic head and neck squamous cell carcinomas. Cancer Radiother. 2020, 24, 559–566. [Google Scholar] [CrossRef]
- You, G.R.; Chang, J.T.; Li, Y.L.; Chen, Y.J.; Huang, Y.C.; Fan, K.H.; Chen, Y.C.; Kang, C.J.; Cheng, A.J. Molecular interplays between cell invasion and radioresistance that lead to poor prognosis in head-neck cancer. Front. Oncol. 2021, 11, 681717. [Google Scholar] [CrossRef]
- Kokame, K.; Kato, H.; Miyata, T. Homocysteine-respondent genes in vascular endothelial cells identified by differential display analysis. GRP78/BiP and novel genes. J. Biol. Chem. 1996, 271, 29659–29665. [Google Scholar] [CrossRef] [Green Version]
- Zhou, D.; Salnikow, K.; Costa, M. Cap43, a novel gene specifically induced by Ni2+ compounds. Cancer Res. 1998, 58, 2182–2189. [Google Scholar]
- Kurdistani, S.K.; Arizti, P.; Reimer, C.L.; Sugrue, M.M.; Aaronson, S.A.; Lee, S.W. Inhibition of tumor cell growth by RTP/rit42 and its responsiveness to p53 and DNA damage. Cancer Res. 1998, 58, 4439–4444. [Google Scholar]
- Agarwala, K.L.; Kokame, K.; Kato, H.; Miyata, T. Phosphorylation of RTP, an ER stress-responsive cytoplasmic protein. Biochem. Biophys. Res. Commun. 2000, 272, 641–647. [Google Scholar] [CrossRef]
- Lachat, P.; Shaw, P.; Gebhard, S.; van Belzen, N.; Chaubert, P.; Bosman, F.T. Expression of NDRG1, a differentiation-related gene, in human tissues. Histochem. Cell Biol. 2002, 118, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Kitowska, A.; Pawelczyk, T. N-myc downstream regulated 1 gene and its place in the cellular machinery. Acta Biochim. Pol. 2010, 57, 15–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croessmann, S.; Wong, H.Y.; Zabransky, D.J.; Chu, D.; Mendonca, J.; Sharma, A.; Mohseni, M.; Rosen, D.M.; Scharpf, R.B.; Cidado, J.; et al. NDRG1 links p53 with proliferation-mediated centrosome homeostasis and genome stability. Proc. Natl. Acad. Sci. USA 2015, 112, 11583–11588. [Google Scholar] [CrossRef] [Green Version]
- Sevinsky, C.J.; Khan, F.; Kokabee, L.; Darehshouri, A.; Maddipati, K.R.; Conklin, D.S. NDRG1 regulates neutral lipid metabolism in breast cancer cells. Breast Cancer Res. 2018, 20, 55. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, B.; Hu, Q.; Qin, Y.; Xu, W.; Shi, S.; Liang, C.; Meng, Q.; Xiang, J.; Liang, D.; et al. A new facet of NDRG1 in pancreatic ductal adenocarcinoma: Suppression of glycolytic metabolism. Int. J. Oncol. 2017, 50, 1792–1800. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Feng, B.; Zhu, F.; Yu, C.; Lu, J.; Pan, M.; He, Z.; Wangpu, X.; Sun, J.; Yang, X. N-myc downstream-regulated gene 1 promotes apoptosis in colorectal cancer via up-regulating death receptor 4. Oncotarget 2017, 8, 82593–82608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Zhu, F.; Yu, C.; Lu, J.; Zhang, L.; Lv, Y.; Sun, J.; Zheng, M. N-myc downstream-regulated gene 1 promotes oxaliplatin-triggered apoptosis in colorectal cancer cells via enhancing the ubiquitination of Bcl-2. Oncotarget 2017, 8, 47709–47724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, L.; Liu, K.; Wang, X.; Chen, H.; Zhou, J.; Wu, X.; Liu, T.; Yang, Y.; Yang, X.; Cui, D.; et al. NDRG1 disruption alleviates cisplatin/sodium glycididazole-induced DNA damage response and apoptosis in ERCC1-defective lung cancer cells. Int. J. Biochem. Cell Biol. 2018, 100, 54–60. [Google Scholar] [CrossRef]
- Wang, H.; Li, W.; Xu, J.; Zhang, T.; Zuo, D.; Zhou, Z.; Lin, B.; Wang, G.; Wang, Z.; Sun, W.; et al. NDRG1 inhibition sensitizes osteosarcoma cells to combretastatin A-4 through targeting autophagy. Cell Death Dis. 2017, 8, e3048. [Google Scholar] [CrossRef]
- Merlot, A.M.; Porter, G.M.; Sahni, S.; Lim, E.G.; Peres, P.; Richardson, D.R. The metastasis suppressor, NDRG1, differentially modulates the endoplasmic reticulum stress response. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 2094–2110. [Google Scholar] [CrossRef]
- Li, E.Y.; Huang, W.Y.; Chang, Y.C.; Tsai, M.H.; Chuang, E.Y.; Kuok, Q.Y.; Bai, S.T.; Chao, L.Y.; Sher, Y.P.; Lai, L.C. Aryl Hydrocarbon Receptor Activates NDRG1 Transcription under Hypoxia in Breast Cancer Cells. Sci. Rep. 2016, 6, 20808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiffon, C. Histone Deacetylase Inhibition Restores Expression of Hypoxia-Inducible Protein NDRG1 in Pancreatic Cancer. Pancreas 2018, 47, 200–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cangul, H. Hypoxia upregulates the expression of the NDRG1 gene leading to its overexpression in various human cancers. BMC Genet. 2004, 5, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dos Santos, M.; da Cunha Mercante, A.M.; Nunes, F.D.; Leopoldino, A.M.; de Carvalho, M.B.; Gazito, D.; López, R.V.; Chiappini, P.B.; de Carvalho Neto, P.B.; Fukuyama, E.E.; et al. Prognostic significance of NDRG1 expression in oral and oropharyngeal squamous cell carcinoma. Mol. Biol. Rep. 2012, 39, 10157–10165. [Google Scholar] [CrossRef]
- Ureshino, H.; Murakami, Y.; Watari, K.; Izumi, H.; Kawahara, A.; Kage, M.; Arao, T.; Nishio, K.; Yanagihara, K.; Kinoshita, H.; et al. N-myc downstream regulated gene 1 (NDRG1) promotes metastasis of human scirrhous gastric cancer cells through epithelial mesenchymal transition. PLoS ONE 2012, 7, e41312. [Google Scholar] [CrossRef]
- Li, A.; Zhu, X.; Wang, C.; Yang, S.; Qiao, Y.; Qiao, R.; Zhang, J. Upregulation of NDRG1 predicts poor outcome and facilitates disease progression by influencing the EMT process in bladder cancer. Sci. Rep. 2019, 9, 5166. [Google Scholar] [CrossRef]
- Song, Y.; Lv, L.; Du, J.; Yue, L.; Cao, L. Correlation of N-myc downstream-regulated gene 1 subcellular localization and lymph node metastases of colorectal neoplasms. Biochem. Biophys. Res. Commun. 2013, 439, 241–246. [Google Scholar] [CrossRef] [Green Version]
- Inagaki, Y.; Tang, W.; Xu, H.L.; Guo, Q.; Mafune, K.; Konishi, T.; Nakata, M.; Sugawara, Y.; Kokudo, N. Localization of N-myc downstream-regulated gene 1 in gastric cancer tissue. Dig. Liver Dis. 2009, 41, 96–103. [Google Scholar] [CrossRef]
- Dai, T.; Dai, Y.; Murata, Y.; Husni, R.E.; Nakano, N.; Sakashita, S.; Noguchi, M. The prognostic significance of N-myc downregulated gene 1 in lung adenocarcinoma. Pathol. Int. 2018, 68, 224–231. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, D.; Li, Y.; Yan, S.; Dang, H.; Yue, H.; Ling, J.; Chen, F.; Zhao, Y.; Gou, L.; et al. Long noncoding RNA CCAT2 promotes hepatocellular carcinoma proliferation and metastasis through up-regulation of NDRG1. Exp. Cell Res. 2019, 379, 19–29. [Google Scholar] [CrossRef]
- Chang, X.; Xu, X.; Ma, J.; Xue, X.; Li, Z.; Deng, P.; Zhang, S.; Zhi, Y.; Chen, J.; Dai, D. NDRG1 expression is related to the progression and prognosis of gastric cancer patients through modulating proliferation, invasion and cell cycle of gastric cancer cells. Mol. Biol. Rep. 2014, 41, 6215–6223. [Google Scholar] [CrossRef] [PubMed]
- Ai, R.; Sun, Y.; Guo, Z.; Wei, W.; Zhou, L.; Liu, F.; Hendricks, D.T.; Xu, Y.; Zhao, X. NDRG1 overexpression promotes the progression of esophageal squamous cell carcinoma through modulating Wnt signaling pathway. Cancer Biol. Ther. 2016, 17, 943–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villodre, E.S.; Hu, X.; Eckhardt, B.; Larson, R.; Huo, L.; Yoon, E.C.; Gong, Y.; Song, J.; Liu, S.; Ueno, N.T.; et al. NDRG1 in aggressive breast cancer progression and brain metastasis. J. Natl. Cancer Inst. 2022, 114, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Bracher-Manecke, J.C.; Zhao, X.; Davies, N.H.; Zhou, L.; Ai, Z.; Oliver, L.; Vallette, F.; Hendricks, D.T. Oncogenic but non-essential role of N-myc downstream regulated gene 1 in the progression of esophageal squamous cell carcinoma. Cancer Biol. Ther. 2013, 14, 164–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.F.; Zhou, Y.; Tao, F.; Chai, S.J.; Xu, X.; Yang, Y.; Yang, Y.; Xu, H.; Wang, K. N-myc downstream regulated gene 1 (NDRG1) promotes the stem-like properties of lung cancer cells through stabilized c-Myc. Cancer Lett. 2017, 401, 53–62. [Google Scholar] [CrossRef]
- Luo, Q.; Wang, C.Q.; Yang, L.Y.; Gao, X.M.; Sun, H.T.; Zhang, Y.; Zhang, K.L.; Zhu, Y.; Zheng, Y.; Sheng, Y.Y.; et al. FOXQ1/NDRG1 axis exacerbates hepatocellular carcinoma initiation via enhancing crosstalk between fibroblasts and tumor cells. Cancer Lett. 2018, 417, 21–34. [Google Scholar] [CrossRef]
- Narayanan, A.; Gagliardi, F.; Gallotti, A.L.; Mazzoleni, S.; Cominelli, M.; Fagnocchi, L.; Pala, M.; Piras, I.S.; Zordan, P.; Moretta, N.; et al. The proneural gene ASCL1 governs the transcriptional subgroup affiliation in glioblastoma stem cells by directly repressing the mesenchymal gene NDRG1. Cell Death Differ. 2019, 26, 1813–1831. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.C.; Shin, Y.K.; Kim, Y.A.; Jang, S.G.; Ku, J.L. Identification of genes inducing resistance to ionizing radiation in human rectal cancer cell lines: Re-sensitization of radio-resistant rectal cancer cells through down regulating NDRG1. BMC Cancer 2018, 18, 594. [Google Scholar] [CrossRef]
- Zhang, D.; Jia, J.; Zhao, G.; Yue, M.; Yang, H.Y.; Wang, J.X. NDRG1 promotes the multidrug resistance of neuroblastoma cells with upregulated expression of drug resistant proteins. Biomed. Pharmacother. 2015, 76, 46–51. [Google Scholar] [CrossRef]
- Du, A.L.; Jiang, Y.F.; Fan, C.F. NDRG1 downregulates ATF3 and inhibits cisplatin-induced cytotoxicity in lung cancer A549 Cells. Int. J. Med. Sci. 2018, 15, 1502–1507. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.S.; Sun, W.; Sun, M.X.; Fu, Z.Z.; Zhou, C.H.; Wang, C.R.; Zuo, D.; Zhou, Z.; Wang, G.; Zhang, T.; et al. HER4 promotes cell survival and chemoresistance in osteosarcoma via interaction with NDRG1. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1839–1849. [Google Scholar] [CrossRef] [PubMed]
- Ueki, S.; Fujishima, F.; Kumagai, T.; Ishida, H.; Okamoto, H.; Takaya, K.; Sato, C.; Taniyma, Y.; Kamei, T.; Sasano, H. GR, Sgk1, and NDRG1 in esophageal squamous cell carcinoma: Their correlation with therapeutic outcome of neoadjuvant chemotherapy. BMC Cancer 2020, 20, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angst, E.; Dawson, D.W.; Stroka, D.; Gloor, B.; Park, J.; Candinas, D.; Reber, H.A.; Hines, O.J.; Eibl, G. N-myc downstream regulated gene-1 expression correlates with reduced pancreatic cancer growth and increased apoptosis in vitro and in vivo. Surgery 2011, 149, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Pan, P.; Qiao, P.F.; Liu, R.L. Downregulation of N-myc downstream regulated gene 1 caused by the methylation of CpG islands of NDRG1 promoter promotes proliferation and invasion of prostate cancer cells. Int. J. Oncol. 2015, 47, 1001–1008. [Google Scholar] [CrossRef]
- Lv, X.H.; Chen, J.W.; Zhao, G.; Feng, Z.Z.; Yang, D.H.; Sun, W.W.; Fan, J.S.; Zhu, G.H. N-myc downstream-regulated gene 1/Cap43 may function as tumor suppressor in endometrial cancer. J. Cancer Res. Clin. 2012, 138, 1703–1715. [Google Scholar] [CrossRef]
- Liu, W.; Iiizumi-Gairani, M.; Okuda, H.; Kobayashi, A.; Watabe, M.; Pai, S.K.; Pandey, P.R.; Xing, F.; Fukuda, K.; Modur, V.; et al. KAI1 gene is engaged in NDRG1 gene-mediated metastasis suppression through the ATF3-NF kappa B complex in human prostate cancer. J. Biol. Chem. 2011, 286, 18949–18959. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.L.; Gao, Q.L.; Zeng, S.; Shen, H. Knockdown of NDRG1 promote epithelial-mesenchymal transition of colorectal cancer via NF-B signaling. J. Surg. Oncol. 2016, 114, 520–527. [Google Scholar] [CrossRef]
- Hu, Z.Y.; Xie, W.B.; Yang, F.; Xiao, L.W.; Wang, X.Y.; Chen, S.Y.; Li, Z.G. NDRG1 attenuates epithelial-mesenchymal transition of nasopharyngeal cancer cells via blocking Smad2 signaling. Biochim. Biophys. Acta Mol. Basis Dis. 2015, 1852, 1876–1886. [Google Scholar] [CrossRef] [Green Version]
- Chang, X.J.; Xu, X.Y.; Xue, X.Y.; Ma, J.G.; Li, Z.H.; Deng, P.; Chen, J.; Zhang, S.; Zhi, Y.; Dai, D. NDRG1 controls gastric cancer migration and invasion through regulating MMP-9. Pathol. Oncol. Res. 2016, 22, 789–796. [Google Scholar] [CrossRef]
- Lee, J.C.; Chung, L.C.; Chen, Y.J.; Feng, T.H.; Juang, H.H. N-myc downstream-regulated gene 1 downregulates cell proliferation, invasiveness, and tumorigenesis in human oral squamous cell carcinoma. Cancer Lett. 2014, 355, 242–252. [Google Scholar] [CrossRef]
- Cen, G.; Zhang, K.D.; Cao, J.; Qiu, Z.J. Downregulation of the N-myc downstream regulated gene 1 is related to enhanced proliferation, invasion and migration of pancreatic cancer. Oncol. Rep. 2017, 37, 1189–1195. [Google Scholar] [CrossRef] [PubMed]
- Wangpu, X.Z.; Yang, X.; Zhao, J.K.; Lu, J.Y.; Guan, S.P.; Lu, J.; Kovacevic, Z.; Liu, W.; Mi, L.; Jin, R.; et al. The metastasis suppressor, NDRG1, inhibits “stemness” of colorectal cancer via down-regulation of nuclear beta-catenin and CD44. Oncotarget 2015, 6, 33893–33911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, S.J.; You, G.R.; Chang, J.T.; Cheng, A.J. Systemic analysis and identification of dysregulated panel lncRNAs contributing to poor prognosis in head-neck cancer. Front. Oncol. 2021, 11, 731752. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Chang, J.T.; Chiu, C.; Lu, Y.C.; Li, Y.L.; Chiang, C.H.; You, G.R.; Lee, L.Y.; Cheng, A.J. Areca nut contributes to oral malignancy through facilitating the conversion of cancer stem cells. Mol. Carcinogen. 2016, 55, 1012–1023. [Google Scholar] [CrossRef]
- Li, Y.L.; Chang, J.T.; Lee, L.Y.; Fan, K.H.; Lu, Y.C.; Li, Y.C.; Chiang, C.H.; You, G.R.; Chen, H.Y.; Cheng, A.J. GDF15 contributes to radioresistance and cancer stemness of head and neck cancer by regulating cellular reactive oxygen species via a SMAD-associated signaling pathway. Oncotarget 2017, 8, 1508–1528. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.C.; Cheng, A.J.; Lee, L.Y.; You, G.R.; Li, Y.L.; Chen, H.Y.; Chang, J.T. MiR-520b as a novel molecular target for suppressing stemness phenotype of head-neck cancer by inhibiting CD44. Sci. Rep. 2017, 7, 2042. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.Y.; Chang, J.T.C.; Chien, K.Y.; Lee, Y.S.; You, G.R.; Cheng, A.J. The endogenous GRP78 interactome in human head and neck cancers: A deterministic role of cell surface GRP78 in cancer stemness. Sci. Rep. 2018, 8, 536. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.J.; Lee, L.Y.; Chao, Y.K.; Chang, J.T.; Lu, Y.C.; Li, H.F.; Chiu, C.C.; Li, Y.C.; Li, Y.L.; Chiou, J.F.; et al. DSG3 facilitates cancer cell growth and invasion through the DSG3-Plakoglobin-TCF/LEF-Myc/Cyclin D1/MMP signaling pathway. PLoS ONE 2013, 8, e64088. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.T.C.; Yang, H.T.; Wang, T.C.V.; Cheng, A.J. Upstream stimulatory factor (USF) as a transcriptional suppressor of human telomerase reverse transcriptase (hTERT) in oral cancer cells. Mol. Carcinogen. 2005, 44, 183–192. [Google Scholar] [CrossRef]
- Schulz, A.; Meyer, F.; Dubrovska, A.; Borgmann, K. Cancer stem cells and radioresistance: DNA repair and beyond. Cancers 2019, 11, 862. [Google Scholar] [CrossRef] [Green Version]
- Kanno, Y.; Chen, C.Y.; Lee, H.L.; Chiou, J.F.; Chen, Y.J. Molecular Mechanisms of Chemotherapy Resistance in Head and Neck Cancers. Front. Oncol. 2021, 11, 640392. [Google Scholar] [CrossRef] [PubMed]
- Aubol, B.E.; Nolen, B.; Shaffer, J.; Ghosh, G.; Adams, J.A. Novel destabilization of nucleotide binding by the gamma phosphate of ATP in the yeast SR protein kinase Sky1p. Biochemestry 2003, 42, 12813–12820. [Google Scholar] [CrossRef] [PubMed]
- Caceres, J.F.; Screaton, G.R.; Krainer, A.R. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Gene Dev. 1998, 12, 55–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, M.C.; Lin, R.I.; Tarn, W.Y. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 10154–10159. [Google Scholar] [CrossRef] [Green Version]
- Murray, J.T.; Campbell, D.G.; Morrice, N.; Auld, G.C.; Shpiro, N.; Marquez, R.; Peggie, M.; Bain, J.; Bloomberg, G.B.; Grahammer, F.; et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem. J. 2004, 384, 477–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, E.; McCue, L.A.; Lawrence, C.E.; Dordick, J.S. Identification of a novel class in the alpha/beta hydrolase fold superfamily: The N-myc differentiation-related proteins. Proteins 2002, 47, 163–168. [Google Scholar] [CrossRef]
- Qu, X.H.; Zhai, Y.; Wei, H.D.; Zhang, C.G.; Xing, G.C.; Yu, Y.T.; He, F. Characterization and expression of three novel differentiation-related genes belong to the human NDRG gene family. Mol. Cell Biochem. 2002, 229, 35–44. [Google Scholar] [CrossRef]
- Zhou, R.H.; Kokame, K.; Tsukamoto, Y.; Yutani, C.; Kato, H.; Miyata, T. Characterization of the human NDRG gene family: A newly identified member, NDRG4, is specifically expressed in brain and heart. Genomics 2001, 73, 86–97. [Google Scholar] [CrossRef]
- Sugiki, T.; Murakami, M.; Taketomi, Y.; Kikuchi-Yanoshita, R.; Kudo, I. N-myc downregulated gene 1 is a phosphorylated protein in mast cells. Biol. Pharm. Bull. 2004, 27, 624–627. [Google Scholar] [CrossRef] [Green Version]
- Tu, L.C.; Yan, X.W.; Hood, L.; Lin, B.Y. Proteomics analysis of the interactome of N-myc downstream regulated gene 1 and its interactions with the androgen response program in prostate cancer cells. Mol. Cell Proteom. 2007, 6, 575–588. [Google Scholar] [CrossRef] [Green Version]
- Drake, J.M.; Paull, E.; Graham, N.A.; Lee, J.K.; Smith, B.A.; Titz, B.; Stoyanova, T.; Faltermeier, C.M.; Uzunangelov, V.; Carlin, D.E.; et al. Phosphoproteome integration reveals patient-specific networks in prostate cancer. Cell 2016, 166, 1041–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, K.C.; Menezes, S.V.; Kalinowski, D.S.; Sahni, S.; Jansson, P.J.; Kovacevic, Z.; Richardson, D.R. Identification of differential phosphorylation and sub-cellular localization of the metastasis suppressor, NDRG1. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 2644–2663. [Google Scholar] [CrossRef] [PubMed]
- McCaig, C.; Potter, L.; Abramczyk, O.; Murray, J.T. Phosphorylation of NDRG1 is temporally and spatially controlled during the cell cycle. Biochem. Biophys. Res. Commun. 2011, 411, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Hosoi, F.; Izumi, H.; Maruyama, Y.; Ureshino, H.; Watari, K.; Kohno, K.; Kuwano, M.; Ono, M. Identification of sites subjected to serine/threonine phosphorylation by SGK1 affecting N-myc downstream-regulated gene 1 (NDRG1)/Cap43-dependent suppression of angiogenic CXC chemokine expression in human pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2010, 396, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Miyayama, T.; Matsuoka, M. Increased expression and activation of serum- and glucocorticoid-inducible kinase-1 (SGK1) by cadmium in HK-2 renal proximal tubular epithelial cells. Environ. Toxicol. Pharmacol. 2014, 38, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Miyata, S.; Koyama, Y.; Takemoto, K.; Yoshikawa, K.; Ishikawa, T.; Taniguchi, M.; Inoue, K.; Aoki, M.; Hori, O.; Katayama, T.; et al. Plasma Corticosterone Activates SGK1 and Induces Morphological Changes in Oligodendrocytes in Corpus Callosum. PLoS ONE 2011, 6, e19859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarrinpashneh, E.; Poggioli, T.; Sarathchandra, P.; Lexow, J.; Monassier, L.; Terracciano, C.; Lang, F.; Damilano, F.; Zhou, J.Q.; Rosenzweig, A.; et al. Ablation of SGK1 impairs endothelial cell migration and tube formation leading to decreased neo-angiogenesis following myocardial infarction. PLoS ONE 2013, 8, e80268. [Google Scholar] [CrossRef] [Green Version]
- Ledet, R.J.; Ruff, S.E.; Wang, Y.; Nayak, S.; Schneider, J.A.; Ueberheide, B.; Logan, S.K.; Garabedian, M.J. Identification of PIM1 substrates reveals a role for NDRG1 phosphorylation in prostate cancer cellular migration and invasion. Commun. Biol. 2021, 4, 36. [Google Scholar] [CrossRef]
- Lu, W.J.; Chua, M.S.; Wei, W.; So, S.K. NDRG1 promotes growth of hepatocellular carcinoma cells by directly interacting with GSK-3 beta and Nur77 to prevent beta-catenin degradation. Oncotarget 2015, 6, 29847–29859. [Google Scholar] [CrossRef]
- de Lima, J.M.; Morand, G.B.; Macedo, C.C.S.; Diesel, L.; Hier, M.P.; Mlynarek, A.; Kowalski, L.P.; Maschietto, M.; Alaoui-Jamali, M.A.; da Silva, S.D. NDRG1 deficiency is associated with regional metastasis in oral cancer by inducing epithelial-mesenchymal transition. Carcinogenesis 2020, 41, 769–777. [Google Scholar] [CrossRef]
- Dickinson, A.; Saraswat, M.; Mäkitie, A.; Silén, R.; Hagström, J.; Haglund, C.; Joenväärä, S.; Silén, S. Label-free tissue proteomics can classify oral squamous cell carcinoma from healthy tissue in a stage-specific manner. Oral Oncol. 2018, 86, 206–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]












Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
You, G.-R.; Chang, J.T.; Li, H.-F.; Cheng, A.-J. Multifaceted and Intricate Oncogenic Mechanisms of NDRG1 in Head and Neck Cancer Depend on Its C-Terminal 3R-Motif. Cells 2022, 11, 1581. https://0-doi-org.brum.beds.ac.uk/10.3390/cells11091581
You G-R, Chang JT, Li H-F, Cheng A-J. Multifaceted and Intricate Oncogenic Mechanisms of NDRG1 in Head and Neck Cancer Depend on Its C-Terminal 3R-Motif. Cells. 2022; 11(9):1581. https://0-doi-org.brum.beds.ac.uk/10.3390/cells11091581
Chicago/Turabian StyleYou, Guo-Rung, Joseph T. Chang, Hsiao-Fan Li, and Ann-Joy Cheng. 2022. "Multifaceted and Intricate Oncogenic Mechanisms of NDRG1 in Head and Neck Cancer Depend on Its C-Terminal 3R-Motif" Cells 11, no. 9: 1581. https://0-doi-org.brum.beds.ac.uk/10.3390/cells11091581