Tracing Early Neurodevelopment in Schizophrenia with Induced Pluripotent Stem Cells
Abstract
:1. Introduction
1.1. The Neurodevelopmental Hypothesis of SCZ
1.2. The Genetic Architecture of SCZ
1.3. Early Brain Development
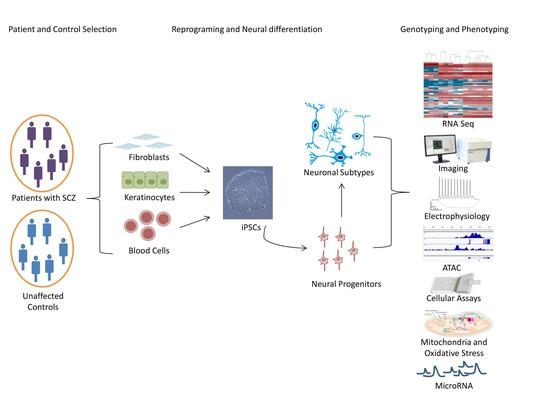
1.4. The Rational of iPSC-Based Disease Modeling
1.5. Tracing Early Neurodevelopment in SCZ
1.5.1. Neurodevelopment and Differentiation of SCZ iPSC-Forebrain Neurons
1.5.2. Dopaminergic Neurons
1.5.3. Hippocampus
1.5.4. Oxidative Stress in SCZ iPSCs
1.5.5. MicroRNAs in SCZ iPSCs
2. Future Directions
2.1. High Resolution Karyotype Maps
2.2. Assessing Differentiation Capacity
2.3. Modeling of Polygenic Disorders
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization Mental Disorders Fact Sheet Schizophrenia 2016. Available online: http://www.who.int/mediacentre/factsheets/fs397/en/ (accessed on 2 August 2018).
- Olfson, M.; Gerhard, T.; Huang, C.; Crystal, S.; Stroup, T.S. Premature Mortality among Adults with Schizophrenia in the United States. JAMA Psychiatry 2015, 72, 1172–1181. [Google Scholar] [CrossRef] [PubMed]
- Thornicroft, G. Physical health disparities and mental illness: The scandal of premature mortality. Br. J. Psychiatry 2011, 199, 441–442. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Mental Disorders Fact Sheet 2017. Available online: http://www.who.int/mediacentre/factsheets/fs396/en/ (accessed on 2 August 2018).
- Insel, T.R. Rethinking schizophrenia. Nature 2010, 468, 187–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owen, M.J.; O’Donovan, M.C. Schizophrenia and the neurodevelopmental continuum: Evidence from genomics. World Psychiatry 2017, 16, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moskowitz, A.; Heim, G. Eugen Bleuler’s Dementia praecox or the group of schizophrenias (1911): A centenary appreciation and reconsideration. Schizophr Bull. 2011, 37, 471–479. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. The International Pilot Study of Schizophrenia 1973. Available online: http://www.who.int/iris/handle/10665/39405 (accessed on 10 August 2018).
- Mueser, K.T.; Jeste, D.V. Clinical Handbook of Schizophrenia; Guilford Press: New York, NY, USA, 2008; ISBN 978-1-60623-045-9. [Google Scholar]
- NHS Choices Schizophrenia—Symptoms. 2018. Available online: https://www.nhs.uk/conditions/schizophrenia/symptoms/ (accessed on 14 August 2018).
- Willner, P. The dopamine hypothesis of schizophrenia: Current status, future prospects. Int. Clin. Psychopharmacol. 1997, 12, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.G.; Woerner, M.G.; McMeniman, M.; Mendelowitz, A.; Bilder, R.M. Symptomatic and functional recovery from a first episode of schizophrenia or schizoaffective disorder. Am. J. Psychiatry 2004, 161, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Javitt, D.C. Glutamatergic theories of schizophrenia. Isr. J. Psychiatry Relat. Sci. 2010, 47, 4–16. [Google Scholar] [PubMed]
- Jablensky, A.; McNeil, T.F.; Morgan, V.A. Barbara Fish and a Short History of the Neurodevelopmental Hypothesis of Schizophrenia. Schizophr. Bull. 2017, 43, 1158–1163. [Google Scholar] [CrossRef] [PubMed]
- Fish, B. The detection of schizophrenia in infancy; a preliminary report. J. Nerv. Ment. Dis. 1957, 125, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.W.; Murray, R.M. Obstetric complications, neurodevelopmental deviance, and risk of schizophrenia. J. Psychiatr. Res. 1987, 21, 413–421. [Google Scholar] [CrossRef]
- Murray, R.M.; Lewis, S.W. Is schizophrenia a neurodevelopmental disorder? Br. Med. J. (Clin. Res. Ed.) 1987, 295, 681–682. [Google Scholar] [CrossRef] [Green Version]
- Weinberger, D.R. Implications of normal brain development for the pathogenesis of schizophrenia. Arch. Gen. Psychiatry 1987, 44, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Kneeland, R.E.; Fatemi, S.H. Viral infection, inflammation and schizophrenia. Prog. Neuropsychopharmacol. Biol. Psychiatry 2013, 42, 35–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellman, L.M.; Deicken, R.F.; Vinogradov, S.; Kremen, W.S.; Poole, J.H.; Kern, D.M.; Tsai, W.Y.; Schaefer, C.A.; Brown, A.S. Structural brain alterations in schizophrenia following fetal exposure to the inflammatory cytokine interleukin-8. Schizophr. Res. 2010, 121, 46–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannon, M.; Jones, P.B.; Murray, R.M. Obstetric complications and schizophrenia: Historical and meta-analytic review. Am. J. Psychiatry 2002, 159, 1080–1092. [Google Scholar] [CrossRef] [PubMed]
- St Clair, D.; Xu, M.; Wang, P.; Yu, Y.; Fang, Y.; Zhang, F.; Zheng, X.; Gu, N.; Feng, G.; Sham, P.; et al. Rates of adult schizophrenia following prenatal exposure to the Chinese famine of 1959–1961. JAMA 2005, 294, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Susser, E.S.; Lin, S.P. Schizophrenia after prenatal exposure to the Dutch Hunger Winter of 1944–1945. Arch. Gen. Psychiatry 1992, 49, 983–988. [Google Scholar] [CrossRef] [PubMed]
- Cannon, T.D.; Bearden, C.E.; Hollister, J.M.; Rosso, I.M.; Sanchez, L.E.; Hadley, T. Childhood cognitive functioning in schizophrenia patients and their unaffected siblings: A prospective cohort study. Schizophr. Bull. 2000, 26, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Cornblatt, B.; Obuchowski, M.; Roberts, S.; Pollack, S.; Erlenmeyer-Kimling, L. Cognitive and behavioral precursors of schizophrenia. Dev. Psychopathol. 1999, 11, 487–508. [Google Scholar] [CrossRef] [PubMed]
- Erlenmeyer-Kimling, L.; Rock, D.; Roberts, S.A.; Janal, M.; Kestenbaum, C.; Cornblatt, B.; Adamo, U.H.; Gottesman, I.I. Attention, memory, and motor skills as childhood predictors of schizophrenia-related psychoses: The New York High-Risk Project. Am. J. Psychiatry 2000, 157, 1416–1422. [Google Scholar] [CrossRef] [PubMed]
- Karayiorgou, M.; Simon, T.J.; Gogos, J.A. 22q11.2 microdeletions: Linking DNA structural variation to brain dysfunction and schizophrenia. Nat. Rev. Neurosci. 2010, 11, 402–416. [Google Scholar] [CrossRef] [PubMed]
- Reichenberg, A.; Caspi, A.; Harrington, H.; Houts, R.; Keefe, R.S.E.; Murray, R.M.; Poulton, R.; Moffitt, T.E. Static and dynamic cognitive deficits in childhood preceding adult schizophrenia: A 30-year study. Am. J. Psychiatry 2010, 167, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, H.J.; Mortensen, E.L.; Schiffman, J.; Reinisch, J.M.; Maeda, J.; Mednick, S.A. Early developmental milestones and risk of schizophrenia: A 45-year follow-up of the Copenhagen Perinatal Cohort. Schizophr. Res. 2010, 118, 41–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodberry, K.A.; Giuliano, A.J.; Seidman, L.J. Premorbid IQ in schizophrenia: A meta-analytic review. Am. J. Psychiatry 2008, 165, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Adriano, F.; Caltagirone, C.; Spalletta, G. Hippocampal volume reduction in first-episode and chronic schizophrenia: A review and meta-analysis. Neuroscientist 2012, 18, 180–200. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, E.C.; Crow, T.J.; Frith, C.D.; Husband, J.; Kreel, L. Cerebral ventricular size and cognitive impairment in chronic schizophrenia. Lancet 1976, 2, 924–926. [Google Scholar] [CrossRef]
- Weinberger, D.R.; Radulescu, E. Finding the Elusive Psychiatric “Lesion” With 21st-Century Neuroanatomy: A Note of Caution. Am. J. Psychiatry 2016, 173, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Wright, I.C.; Rabe-Hesketh, S.; Woodruff, P.W.; David, A.S.; Murray, R.M.; Bullmore, E.T. Meta-analysis of regional brain volumes in schizophrenia. Am. J. Psychiatry 2000, 157, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.D.; Saykin, A.J.; Flashman, L.A.; Riordan, H.J. Hippocampal volume reduction in schizophrenia as assessed by magnetic resonance imaging: A meta-analytic study. Arch. Gen. Psychiatry 1998, 55, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Ward, K.E.; Friedman, L.; Wise, A.; Schulz, S.C. Meta-analysis of brain and cranial size in schizophrenia. Schizophr. Res. 1996, 22, 197–213. [Google Scholar] [CrossRef]
- Lawrie, S.M.; Abukmeil, S.S. Brain abnormality in schizophrenia. A systematic and quantitative review of volumetric magnetic resonance imaging studies. Br. J. Psychiatry 1998, 172, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Suddath, R.L.; Christison, G.W.; Torrey, E.F.; Casanova, M.F.; Weinberger, D.R. Anatomical abnormalities in the brains of monozygotic twins discordant for schizophrenia. N. Engl. J. Med. 1990, 322, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.J. Brains at risk of schizophrenia. Lancet 1999, 353, 3–4. [Google Scholar] [CrossRef]
- Lieberman, J.A. Is schizophrenia a neurodegenerative disorder? A clinical and neurobiological perspective. Biol. Psychiatry 1999, 46, 729–739. [Google Scholar] [CrossRef]
- Harrison, P.J. Postmortem studies in schizophrenia. Dialogues Clin. Neurosci. 2000, 2, 349–357. [Google Scholar] [PubMed]
- Bakhshi, K.; Chance, S.A. The neuropathology of schizophrenia: A selective review of past studies and emerging themes in brain structure and cytoarchitecture. Neuroscience 2015, 303, 82–102. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstein, P.; Yip, B.H.; Björk, C.; Pawitan, Y.; Cannon, T.D.; Sullivan, P.F.; Hultman, C.M. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: A population-based study. Lancet 2009, 373, 234–239. [Google Scholar] [CrossRef]
- Wray, N.R.; Gottesman, I.I. Using summary data from the danish national registers to estimate heritabilities for schizophrenia, bipolar disorder, and major depressive disorder. Front. Genet. 2012, 3, 118. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, P.F.; Kendler, K.S.; Neale, M.C. Schizophrenia as a complex trait: Evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 2003, 60, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, P.F.; Daly, M.J.; O’Donovan, M. Genetic architectures of psychiatric disorders: The emerging picture and its implications. Nat. Rev. Genet. 2012, 13, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.M.; Moran, J.L.; Fromer, M.; Ruderfer, D.; Solovieff, N.; Roussos, P.; O’Dushlaine, C.; Chambert, K.; Bergen, S.E.; Kähler, A.; et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 2014, 506, 185–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ripke, S.; Neale, B.M.; Corvin, A.; Walters, J.T.; Farh, K.H.; Holmans, P.A.; Lee, P.; Bulik-Sullivan, B.; Collier, D.A.; Huang, H. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar] [CrossRef] [Green Version]
- Finucane, H.; Reshef, Y.; Anttila, V.; Slowikowski, K.; Gusev, A.; Byrnes, A.; Gazal, S.; Loh, P.-R.; Lareau, C.; Shoresh, N.; et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. bioRxiv 2017, 103069. [Google Scholar] [CrossRef] [Green Version]
- Skene, N.G.; Bryois, J.; Bakken, T.E.; Breen, G.; Crowley, J.J.; Gaspar, H.; Giusti-Rodriguez, P.; Hodge, R.D.; Miller, J.A.; Munoz-Manchado, A.; et al. Genetic Identification Of Brain Cell Types Underlying Schizophrenia. bioRxiv 2017. [Google Scholar] [CrossRef]
- Pardiñas, A.F.; Holmans, P.; Pocklington, A.J.; Escott-Price, V.; Ripke, S.; Carrera, N.; Legge, S.E.; Bishop, S.; Cameron, D.; Hamshere, M.L.; et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 2018, 50, 381–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schork, A.J.; Won, H.; Appadurai, V.; Nudel, R.; Gandal, M.; Delaneau, O.; Hougaard, D.; Baekved-Hansen, M.; Bybjerg-Grauholm, J.; Pedersen, M.G. A genome-wide association study for shared risk across major psychiatric disorders in a nation-wide birth cohort implicates fetal neurodevelopment as a key mediator. bioRxiv 2017, 240911. [Google Scholar] [Green Version]
- Freedman, M.L.; Monteiro, A.N.A.; Gayther, S.A.; Coetzee, G.A.; Risch, A.; Plass, C.; Casey, G.; De Biasi, M.; Carlson, C.; Duggan, D.; et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat. Genet. 2011, 43, 513–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forrest, M.P.; Zhang, H.; Moy, W.; McGowan, H.; Leites, C.; Dionisio, L.E.; Xu, Z.; Shi, J.; Sanders, A.R.; Greenleaf, W.J.; et al. Open Chromatin Profiling in hiPSC-Derived Neurons Prioritizes Functional Noncoding Psychiatric Risk Variants and Highlights Neurodevelopmental Loci. Cell Stem Cell 2017, 21, 305.e8–318.e8. [Google Scholar] [CrossRef] [PubMed]
- Fromer, M.; Pocklington, A.J.; Kavanagh, D.H.; Williams, H.J.; Dwyer, S.; Gormley, P.; Georgieva, L.; Rees, E.; Palta, P.; Ruderfer, D.M.; et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 2014, 506, 179–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genovese, G.; Fromer, M.; Stahl, E.A.; Ruderfer, D.M.; Chambert, K.; Landén, M.; Moran, J.L.; Purcell, S.M.; Sklar, P.; Sullivan, P.F.; et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat. Neurosci. 2016, 19, 1433–1441. [Google Scholar] [CrossRef] [PubMed]
- Ripke, S.; O’Dushlaine, C.; Chambert, K.; Moran, J.L.; Kähler, A.K.; Akterin, S.; Bergen, S.E.; Collins, A.L.; Crowley, J.J.; Fromer, M.; et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013, 45, 1150–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Kichaev, G.; Pasaniuc, B. Contrasting the Genetic Architecture of 30 Complex Traits from Summary Association Data. Am. J. Hum. Genet. 2016, 99, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Boyle, E.A.; Li, Y.I.; Pritchard, J.K. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 2017, 169, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Kandel, E.R.; Schwartz, J.H.; Jessel, T.M.; Siegelbaum, S.A.; Hudspeth, A.J.; Mack, S. Principles of Neural Science, 5th ed.; McGraw-Hill Medical: New York, NY, USA; Lisbon, Portugal; London, UK, 2013; ISBN 978-0-07-139011-8. [Google Scholar]
- Molyneaux, B.J.; Arlotta, P.; Menezes, J.R.L.; Macklis, J.D. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 2007, 8, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Lui, J.H.; Hansen, D.V.; Kriegstein, A.R. Development and evolution of the human neocortex. Cell 2011, 146, 18–36. [Google Scholar] [CrossRef] [PubMed]
- Taverna, E.; Götz, M.; Huttner, W.B. The cell biology of neurogenesis: Toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 2014, 30, 465–502. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.A.; González-Burgos, G. Neuroplasticity of neocortical circuits in schizophrenia. Neuropsychopharmacology 2008, 33, 141–165. [Google Scholar] [CrossRef] [PubMed]
- Rakic, P.; Bourgeois, J.P.; Eckenhoff, M.F.; Zecevic, N.; Goldman-Rakic, P.S. Concurrent overproduction of synapses in diverse regions of the primate cerebral cortex. Science 1986, 232, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Turksen, K.; Nagy, A. Induced Pluripotent Stem (iPS) Cells: Methods and Protocols; Methods in molecular biology; Humana Press: New York, NY, USA, 2016; ISBN 978-1-4939-3055-5. [Google Scholar]
- Verma, P.J.; Sumer, H. Cell Reprogramming: Methods and Protocols; Methods in molecular biology; Humana Press: New York, NY, USA, 2015; ISBN 978-1-4939-2847-7. [Google Scholar]
- Paşca, S.P.; Panagiotakos, G.; Dolmetsch, R.E. Generating human neurons in vitro and using them to understand neuropsychiatric disease. Annu. Rev. Neurosci. 2014, 37, 479–501. [Google Scholar] [CrossRef] [PubMed]
- Brennand, K.; Savas, J.N.; Kim, Y.; Tran, N.; Simone, A.; Hashimoto-Torii, K.; Beaumont, K.G.; Kim, H.J.; Topol, A.; Ladran, I.; et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol. Psychiatry 2015, 20, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Mariani, J.; Simonini, M.V.; Palejev, D.; Tomasini, L.; Coppola, G.; Szekely, A.M.; Horvath, T.L.; Vaccarino, F.M. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2012, 109, 12770–12775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholas, C.R.; Chen, J.; Tang, Y.; Southwell, D.G.; Chalmers, N.; Vogt, D.; Arnold, C.M.; Chen, Y.-J.J.; Stanley, E.G.; Elefanty, A.G.; et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell 2013, 12, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Paşca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.-Y.; O’Rourke, N.A.; Nguyen, K.D.; et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 2015, 12, 671–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, J.L.; de la Torre-Ubieta, L.; Tian, Y.; Parikshak, N.N.; Hernández, I.A.; Marchetto, M.C.; Baker, D.K.; Lu, D.; Hinman, C.R.; Lowe, J.K.; et al. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron 2014, 83, 69–86. [Google Scholar] [CrossRef] [PubMed]
- Brennand, K.J.; Simone, A.; Jou, J.; Gelboin-Burkhart, C.; Tran, N.; Sangar, S.; Li, Y.; Mu, Y.; Chen, G.; Yu, D.; et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 2011, 473, 221–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roussos, P.; Guennewig, B.; Kaczorowski, D.C.; Barry, G.; Brennand, K.J. Activity-Dependent Changes in Gene Expression in Schizophrenia Human-Induced Pluripotent Stem Cell Neurons. JAMA Psychiatry 2016, 73, 1180–1188. [Google Scholar] [CrossRef] [PubMed]
- Topol, A.; Zhu, S.; Tran, N.; Simone, A.; Fang, G.; Brennand, K.J. Altered WNT Signaling in Human Induced Pluripotent Stem Cell Neural Progenitor Cells Derived from Four Schizophrenia Patients. Biol. Psychiatry 2015, 78, e29–e34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robicsek, O.; Karry, R.; Petit, I.; Salman-Kesner, N.; Müller, F.-J.; Klein, E.; Aberdam, D.; Ben-Shachar, D. Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Mol. Psychiatry 2013, 18, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- Hook, V.; Brennand, K.J.; Kim, Y.; Toneff, T.; Funkelstein, L.; Lee, K.C.; Ziegler, M.; Gage, F.H. Human iPSC neurons display activity-dependent neurotransmitter secretion: Aberrant catecholamine levels in schizophrenia neurons. Stem Cell Rep. 2014, 3, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Hartley, B.J.; Tran, N.; Ladran, I.; Reggio, K.; Brennand, K.J. Dopaminergic differentiation of schizophrenia hiPSCs. Mol. Psychiatry 2015, 20, 549–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, D.X.; Di Giorgio, F.P.; Yao, J.; Marchetto, M.C.; Brennand, K.; Wright, R.; Mei, A.; McHenry, L.; Lisuk, D.; Grasmick, J.M.; et al. Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Rep. 2014, 2, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Mei, A.; Paquola, A.C.M.; Stern, S.; Bardy, C.; Klug, J.R.; Kim, S.; Neshat, N.; Kim, H.J.; Ku, M.; et al. Efficient Generation of CA3 Neurons from Human Pluripotent Stem Cells Enables Modeling of Hippocampal Connectivity In Vitro. Cell Stem Cell 2018, 22, 684–697. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, B.D.S.; de Moraes Maciel, R.; Galina, A.; da Silveira, M.S.; dos Santos Souza, C.; Drummond, H.; Pozzatto, E.N.; Silva, H.; Chicaybam, L.; Massuda, R.; et al. Altered oxygen metabolism associated to neurogenesis of induced pluripotent stem cells derived from a schizophrenic patient. Cell Transplant. 2012, 21, 1547–1559. [Google Scholar] [CrossRef] [PubMed]
- Robicsek, O.; Ene, H.M.; Karry, R.; Ytzhaki, O.; Asor, E.; McPhie, D.; Cohen, B.M.; Ben-Yehuda, R.; Weiner, I.; Ben-Shachar, D. Isolated Mitochondria Transfer Improves Neuronal Differentiation of Schizophrenia-Derived Induced Pluripotent Stem Cells and Rescues Deficits in a Rat Model of the Disorder. Schizophr. Bull. 2018, 44, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Siegert, S.; Seo, J.; Kwon, E.J.; Rudenko, A.; Cho, S.; Wang, W.; Flood, Z.; Martorell, A.J.; Ericsson, M.; Mungenast, A.E.; et al. The schizophrenia risk gene product miR-137 alters presynaptic plasticity. Nat. Neurosci. 2015, 18, 1008–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Topol, A.; Zhu, S.; Hartley, B.J.; English, J.; Hauberg, M.E.; Tran, N.; Rittenhouse, C.A.; Simone, A.; Ruderfer, D.M.; Johnson, J.; et al. Dysregulation of miRNA-9 in a Subset of Schizophrenia Patient-Derived Neural Progenitor Cells. Cell Rep. 2016, 15, 1024–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narla, S.T.; Lee, Y.-W.; Benson, C.A.; Sarder, P.; Brennand, K.J.; Stachowiak, E.K.; Stachowiak, M.K. Common developmental genome deprogramming in schizophrenia—Role of Integrative Nuclear FGFR1 Signaling (INFS). Schizophr. Res. 2017, 185, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Iannitelli, A.; Quartini, A.; Tirassa, P.; Bersani, G. Schizophrenia and neurogenesis: A stem cell approach. Neurosci. Biobehav. Rev. 2017, 80, 414–442. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, A.; Sandler, V.M.; Toni, N.; Zhao, C.; Gage, F.H. NMDA-receptor-mediated, cell-specific integration of new neurons in adult dentate gyrus. Nature 2006, 442, 929–933. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Herman, P.; Rothman, D.L.; Agarwal, D.; Hyder, F. Evaluating the gray and white matter energy budgets of human brain function. J. Cereb. Blood Flow Metab. 2017. [Google Scholar] [CrossRef] [PubMed]
- Hroudová, J.; Fišar, Z. Control mechanisms in mitochondrial oxidative phosphorylation. Neural Regen. Res. 2013, 8, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Turrens, J.F. Mitochondrial formation of reactive oxygen species. J. Physiol. (Lond.) 2003, 552, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.D.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef] [PubMed]
- Gitto, E.; Pellegrino, S.; Gitto, P.; Barberi, I.; Reiter, R.J. Oxidative stress of the newborn in the pre- and postnatal period and the clinical utility of melatonin. J. Pineal Res. 2009, 46, 128–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrera, E.; Jiménez, R.; Aruoma, O.I.; Hercberg, S.; Sánchez-García, I.; Fraga, C. Aspects of antioxidant foods and supplements in health and disease. Nutr. Rev. 2009, 67 (Suppl. S1), S140–S144. [Google Scholar] [CrossRef] [PubMed]
- Lain, K.Y.; Roberts, J.M. Contemporary concepts of the pathogenesis and management of preeclampsia. JAMA 2002, 287, 3183–3186. [Google Scholar] [CrossRef] [PubMed]
- Redman, C.W.G.; Sargent, I.L. Pre-eclampsia, the placenta and the maternal systemic inflammatory response—A review. Placenta 2003, 24 (Suppl. SA), S21–S27. [Google Scholar] [CrossRef] [PubMed]
- Andreazza, A.C.; Duong, A.; Young, L.T. Bipolar Disorder as a Mitochondrial Disease. Biol. Psychiatry 2018, 83, 720–721. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, C.R.; O’Donovan, S.M.; McCullumsmith, R.E.; Ramsey, A. Defects in Bioenergetic Coupling in Schizophrenia. Biol. Psychiatry 2018, 83, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, B.D.S.; Cardoso, S.C.; Stelling, M.P.; Cadilhe, D.V.; Rehen, S.K. Valproate reverts zinc and potassium imbalance in schizophrenia-derived reprogrammed cells. Schizophr. Res. 2014, 154, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Flippo, K.H.; Strack, S. An emerging role for mitochondrial dynamics in schizophrenia. Schizophr. Res. 2017, 187, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Khacho, M.; Slack, R.S. Mitochondrial dynamics in the regulation of neurogenesis: From development to the adult brain. Dev. Dyn. 2018, 247, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, H.; Yao, Y.; Zhao, T.; Chen, Y.-Y.; Shen, Y.-L.; Wang, L.-L.; Zhu, Y. Stem cell-derived mitochondria transplantation: A novel strategy and the challenges for the treatment of tissue injury. Stem Cell Res. Ther. 2018, 9, 106. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Rajman, M.; Schratt, G. MicroRNAs in neural development: From master regulators to fine-tuners. Development 2017, 144, 2310–2322. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.X.; Crabtree, G.R.; Yoo, A.S. MicroRNAs: Regulators of neuronal fate. Curr. Opin. Cell Biol. 2013, 25, 215–221. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, R.M.; Gururajan, A.; Dinan, T.G.; Kenny, P.J.; Cryan, J.F. All Roads Lead to the miRNome: MiRNAs Have a Central Role in the Molecular Pathophysiology of Psychiatric Disorders. Trends Pharmacol. Sci. 2016, 37, 1029–1044. [Google Scholar] [CrossRef] [PubMed]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet 2013, 381, 1371–1379. [Google Scholar] [CrossRef]
- Kwon, E.; Wang, W.; Tsai, L.-H. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Mol. Psychiatry 2013, 18, 11–12. [Google Scholar] [CrossRef] [PubMed]
- Lett, T.A.; Chakravarty, M.M.; Chakavarty, M.M.; Felsky, D.; Brandl, E.J.; Tiwari, A.K.; Gonçalves, V.F.; Rajji, T.K.; Daskalakis, Z.J.; Meltzer, H.Y.; et al. The genome-wide supported microRNA-137 variant predicts phenotypic heterogeneity within schizophrenia. Mol. Psychiatry 2013, 18, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Sun, G.; Li, S.; Shi, Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 2009, 16, 365–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauberg, M.E.; Roussos, P.; Grove, J.; Børglum, A.D.; Mattheisen, M. Schizophrenia Working Group of the Psychiatric Genomics Consortium Analyzing the Role of MicroRNAs in Schizophrenia in the Context of Common Genetic Risk Variants. JAMA Psychiatry 2016, 73, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Perkins, D.O.; Jeffries, C.D.; Jarskog, L.F.; Thomson, J.M.; Woods, K.; Newman, M.A.; Parker, J.S.; Jin, J.; Hammond, S.M. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007, 8, R27. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.H.; Zeier, Z.; Xi, L.; Lanz, T.A.; Deng, S.; Strathmann, J.; Willoughby, D.; Kenny, P.J.; Elsworth, J.D.; Lawrence, M.S.; et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc. Natl. Acad. Sci. USA 2012, 109, 3125–3130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreau, M.P.; Bruse, S.E.; David-Rus, R.; Buyske, S.; Brzustowicz, L.M. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol. Psychiatry 2011, 69, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Santarelli, D.M.; Beveridge, N.J.; Tooney, P.A.; Cairns, M.J. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol. Psychiatry 2011, 69, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Du, J.; Qi, Y.; Liang, G.; Wang, T.; Li, S.; Xie, S.; Zeshan, B.; Xiao, Z. Aberrant expression of serum miRNAs in schizophrenia. J. Psychiatr. Res. 2012, 46, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Murai, K.; Sun, G.; Ye, P.; Tian, E.; Yang, S.; Cui, Q.; Sun, G.; Trinh, D.; Sun, O.; Hong, T.; et al. The TLX-miR-219 cascade regulates neural stem cell proliferation in neurodevelopment and schizophrenia iPSC model. Nat. Commun 2016, 7, 10965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlaeger, T.M.; Daheron, L.; Brickler, T.R.; Entwisle, S.; Chan, K.; Cianci, A.; DeVine, A.; Ettenger, A.; Fitzgerald, K.; Godfrey, M.; et al. A comparison of non-integrating reprogramming methods. Nat. Biotechnol. 2015, 33, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Kwon, E.M.; Connelly, J.P.; Hansen, N.F.; Donovan, F.X.; Winkler, T.; Davis, B.W.; Alkadi, H.; Chandrasekharappa, S.C.; Dunbar, C.E.; Mullikin, J.C.; et al. iPSCs and fibroblast subclones from the same fibroblast population contain comparable levels of sequence variations. Proc. Natl. Acad. Sci. USA 2017, 114, 1964–1969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banovich, N.E.; Li, Y.I.; Raj, A.; Ward, M.C.; Greenside, P.; Calderon, D.; Tung, P.Y.; Burnett, J.E.; Myrthil, M.; Thomas, S.M.; et al. Impact of regulatory variation across human iPSCs and differentiated cells. Genome Res. 2018, 28, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Burrows, C.K.; Banovich, N.E.; Pavlovic, B.J.; Patterson, K.; Gallego Romero, I.; Pritchard, J.K.; Gilad, Y. Genetic Variation, Not Cell Type of Origin, Underlies the Majority of Identifiable Regulatory Differences in iPSCs. PLoS Genet. 2016, 12, e1005793. [Google Scholar] [CrossRef] [PubMed]
- Carcamo-Orive, I.; Hoffman, G.E.; Cundiff, P.; Beckmann, N.D.; D’Souza, S.L.; Knowles, J.W.; Patel, A.; Papatsenko, D.; Abbasi, F.; Reaven, G.M.; et al. Analysis of Transcriptional Variability in a Large Human iPSC Library Reveals Genetic and Non-genetic Determinants of Heterogeneity. Cell Stem Cell 2017, 20, 518.e9–532.e9. [Google Scholar] [CrossRef] [PubMed]
- Kilpinen, H.; Goncalves, A.; Leha, A.; Afzal, V.; Alasoo, K.; Ashford, S.; Bala, S.; Bensaddek, D.; Casale, F.P.; Culley, O.J.; et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 2017, 546, 370–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouhani, F.; Kumasaka, N.; de Brito, M.C.; Bradley, A.; Vallier, L.; Gaffney, D. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. 2014, 10, e1004432. [Google Scholar] [CrossRef] [PubMed]
- Ziller, M.J.; Gu, H.; Müller, F.; Donaghey, J.; Tsai, L.T.-Y.; Kohlbacher, O.; De Jager, P.L.; Rosen, E.D.; Bennett, D.A.; Bernstein, B.E.; et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013, 500, 477–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paull, D.; Sevilla, A.; Zhou, H.; Hahn, A.K.; Kim, H.; Napolitano, C.; Tsankov, A.; Shang, L.; Krumholz, K.; Jagadeesan, P.; et al. Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells. Nat. Methods 2015, 12, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Bock, C.; Kiskinis, E.; Verstappen, G.; Gu, H.; Boulting, G.; Smith, Z.D.; Ziller, M.; Croft, G.F.; Amoroso, M.W.; Oakley, D.H.; et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 2011, 144, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.-Y.; Weick, J.P.; Yu, J.; Ma, L.-X.; Zhang, X.-Q.; Thomson, J.A.; Zhang, S.-C. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA 2010, 107, 4335–4340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osafune, K.; Caron, L.; Borowiak, M.; Martinez, R.J.; Fitz-Gerald, C.S.; Sato, Y.; Cowan, C.A.; Chien, K.R.; Melton, D.A. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 2008, 26, 313–315. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Xu, J.; Pang, Z.P.; Ge, W.; Kim, K.J.; Blanchi, B.; Chen, C.; Südhof, T.C.; Sun, Y.E. Integrative genomic and functional analyses reveal neuronal subtype differentiation bias in human embryonic stem cell lines. Proc. Natl. Acad. Sci. USA 2007, 104, 13821–13826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyttälä, A.; Moraghebi, R.; Valensisi, C.; Kettunen, J.; Andrus, C.; Pasumarthy, K.K.; Nakanishi, M.; Nishimura, K.; Ohtaka, M.; Weltner, J.; et al. Genetic Variability Overrides the Impact of Parental Cell Type and Determines iPSC Differentiation Potential. Stem Cell Rep. 2016, 6, 200–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, A.; Sportelli, V.; Ziller, M.; Spengler, D. Switch-Like Roles for Polycomb Proteins from Neurodevelopment to Neurodegeneration. Epigenomes 2017, 1, 21. [Google Scholar] [CrossRef]
- Soldner, F.; Jaenisch, R. Medicine. iPSC disease modeling. Science 2012, 338, 1155–1156. [Google Scholar] [CrossRef] [PubMed]
- Schwartzentruber, J.; Foskolou, S.; Kilpinen, H.; Rodrigues, J.; Alasoo, K.; Knights, A.J.; Patel, M.; Goncalves, A.; Ferreira, R.; Benn, C.L.; et al. HIPSCI Consortium Molecular and functional variation in iPSC-derived sensory neurons. Nat. Genet. 2018, 50, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Honey, C.J.; Kötter, R.; Breakspear, M.; Sporns, O. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc. Natl. Acad. Sci. USA 2007, 104, 10240–10245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gollo, L.L.; Roberts, J.A.; Cropley, V.L.; Di Biase, M.A.; Pantelis, C.; Zalesky, A.; Breakspear, M. Fragility and volatility of structural hubs in the human connectome. Nat. Neurosci. 2018, 21, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Quadrato, G.; Arlotta, P. Studying the Brain in a Dish: 3D Cell Culture Models of Human Brain Development and Disease. Curr. Top. Dev. Biol. 2018, 129, 99–122. [Google Scholar] [CrossRef] [PubMed]
- Quadrato, G.; Brown, J.; Arlotta, P. The promises and challenges of human brain organoids as models of neuropsychiatric disease. Nat. Med. 2016, 22, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, S.D.; Stringer, S.; Heine, V.M.; Posthuma, D. Genetically-Informed Patient Selection for iPSC Studies of Complex Diseases May Aid in Reducing Cellular Heterogeneity. Front. Cell. Neurosci. 2017, 11, 164. [Google Scholar] [CrossRef] [PubMed]
- Gandal, M.J.; Leppa, V.; Won, H.; Parikshak, N.N.; Geschwind, D.H. The road to precision psychiatry: Translating genetics into disease mechanisms. Nat. Neurosci. 2016, 19, 1397–1407. [Google Scholar] [CrossRef] [PubMed]
- Yokota, Y.; Ghashghaei, H.T.; Han, C.; Watson, H.; Campbell, K.J.; Anton, E.S. Radial Glial Dependent and Independent Dynamics of Interneuronal Migration in the Developing Cerebral Cortex. PLoS ONE 2007, 2, e794. [Google Scholar] [CrossRef] [PubMed]



| Ref. | Source | Factors | Met | N° | Auth | Karyo | Pluripotency |
|---|---|---|---|---|---|---|---|
| [75] | fibroblast | OKSML | LV | ≥5 | nd | G-B | ICC, Tera |
| [76] | [75] | OKSML | LV | ≥5 | nd | G-B | ICC, Tera |
| [77] | [75] | OKSML | LV | ≥5 | nd | G-B | ICC, Tera |
| [78] | keratinocyte | OKSM | LV | 1 | nd | G-B | ICC, PCR, PluriTest, EB |
| [79] | [75] | OKSML | LV | ≥5 | nd | G-B | ICC, Tera |
| [80] | [75] | OKSML | LV | ≥5 | nd | G-B | ICC, Tera |
| [81] | [75] | OKSML | LV | ≥5 | nd | G-B | ICC, Tera |
| [82] | [75] | OKSML | LV | ≥5 | nd | G-B | ICC, Tera |
| [83] | fibroblast | OKSM | RV | 2 | nd | G-B | ICC, PCR, EB |
| [70] | [75,77] | OKSML | LV | ≥5 | nd | G-B | ICC, Tera |
| [84] | [78] | OKSM | LV | 1 | nd | G-B | ICC, PCR, PluriTest, EB |
| [85] | fibroblast | APM | LV | na | na | na | ICC |
| [55] | fibroblast | OKSM | LV | dns | nd | G-B | ICC, PluriTest, EB |
| [86] | [75] | OKSML | LV | ≥5 | nd | G-B | ICC, Tera |
| fibroblast | OKSM | Sen | 2–3 | nd | G-B | PCR, FACS | |
| [87] | [75] | OKSML | LV | ≥5 | nd | G-B | ICC, Tera |
| Ref. | Study Design | SCZ vs. Controls | Model | Major Cell Type(s) |
|---|---|---|---|---|
| [75] | Familial SCZ | 4 vs. 3 | iPSC | Forebrain, mixed glutamatergic-GABAergic neurons and NPCs |
| [76] | [75] | 4 vs. 3 | iPSC | Forebrain, mixed glutamatergic- GABAergic neurons |
| [77] | [75] | 4 vs. 6 | iPSC | Forebrain NPCs |
| [78] | Paranoid SCZ | 3 vs. 2 | iPSC | Midbrain dopaminergic and forebrain glutamatergic neurons |
| [79] | [75] | 4 vs. 3 | iPSC | Forebrain, mixed glutamatergic-GABAergic neurons |
| [80] | [75] | 4 vs. 3 | iPSC | FOXA2-positive midbrain dopaminergic neurons |
| [81] | [75] | 4 vs. 3 | iPSC | Dentate gyrus-like granule neurons |
| [82] | [75] | 4 vs. 3 | iPSC | CA3 and dentate gyrus neurons |
| [83] | CR-SCZ | 1 vs. 1 | iPSC | Retinoic acid-induced NPCs |
| [70] | [75] | 4 vs. 6 | iPSC | Forebrain NPCs |
| [84] | [78] | 1 vs. 1 | iPSC | Forebrain glutamatergic neurons |
| [85] | MIR137 risk alleles | 3 mar vs. 3 mir | iNLC | Early neuron-like cells FACS-purified |
| [55] | MIR137 risk SNP | 2 vs. 1 vs. 2 ig | iPSC | Induced forebrain glutamatergic neurons |
| [86] | [75] | 4 vs. 6 | iPSC | Forebrain NPCs |
| Childhood-onset SCZ | 10 vs. 10 | iPSC | Forebrain NPCs | |
| [87] | [75] | 3 vs. 3 | iPSC | Neuron committed forebrain NPCs |
| Ref. | Neural Induction | Patterning/Neural Progenitor Cells → Neural Cells |
|---|---|---|
| [75] | EB-/rosette formation | N2, B27-RA, FGF2 → N2, B27-RA, BDNF, GDNF, cAMP, AA |
| [76] | EB-/rosette formation | N2, B27-RA, FGF2 → N2, B27-RA, BDNF, GDNF, cAMP, AA |
| [77] | EB-/rosette formation | N2, B27-RA, FGF2 |
| [78] | Nog, SB431542 | SHH → SHH, FGF8, BDNF, AA |
| [79] | EB-/rosette formation | N2, B27-RA, FGF2 → N2, B27-RA, BDNF, GDNF, cAMP, AA |
| [80] | LDN193189, SB431542 | SHH8 + FGF8 → BDNF, GDNF, cAMP, AA SHH8 + FGF8, CHIR99021 → BDNF, GDNF, cAMP, AA |
| [81] | DKK1, SB431542, Nog, Cyc | N2, B27, FGF2 → N2, B27, BDNF, Wnt3a, cAMP, AA |
| [82] | DKK1, SB431542, Nog, Cyc | N2, B27, FGF2 → N2, B27, BDNF, Wnt3a, cAMP, AA |
| [83] | Nog, bFGF | RA → FGF2 |
| [70] | EB-/rosette formation | N2, B27-RA, FGF2 |
| [84] | Nog, SB431542 | SHH → SHH, FGF8, BDNF, AA |
| [85] | Lentiviral transduction | N2, bFGF |
| [55] | Dorsomorphin or Nog, SB431542 | N2, B27 or B27-RA, FGF2 |
| [86] | LDN193189, SB431542 | N2, B27-RA, FGF2 |
| [87] | EB-/rosette formation | N2, B27-RA, FGF2 → N2, B27-RA, BDNF, GDNF, cAMP, AA |
| Ref. | Major Methods | Major Findings in SCZ iPSCs Derived Cells |
|---|---|---|
| [75] | Rabies virus transport, electrophysiology, calcium transients, microarray | Reduced neuronal connectivity, maintained synaptic function, altered gene expression in glutamate, cAMP, BMP, and WNT pathways; loxapine application normalizes alterations |
| [76] | Potassium-induced depolarization, RNA-seq | Reduced activity-dependent transcription, coexpression modules are enriched for GWAS risk variants |
| [77] | RNA-seq, reporter assay | Differentially expressed genes from WNT, SHH, BMP, and G-protein coupled signaling |
| [78] | Cell imaging, HPLC, mitochondrial assays | Impaired dopaminergic differentiation and glutamatergic maturation, mitochondria show reduced membrane potential, respiration, and connectivity, and uneven network structure |
| [79] | Potassium-induced depol-arization, HPLC, ICC | Increased basal and activity-dependent cortical secretion of dopamine, epinephrine, and norepinephrine |
| [80] | ICC | SCZ/control-iPSCs differentiate equally well into FOXA2-positive midbrain dopaminergic neurons |
| [81] | WCPC, calcium transients | Delayed differentiation of NPCs into DG-like neurons, reduced spontaneous neurotransmitter release |
| [82] | RNA-seq, SCS, WCPC, MEA | Reduced spontaneous spike and network bursts in mature, but not in early, SCZ iPSC-DG-CA3 circuits |
| [83] | Oxygen consumption, ROS production | Increased extra-mitochondrial oxygen consumption and ROS production that is normalized by valproate |
| [70] | Microarray, mass spectrometry | Abnormal gene and protein expression related to cytoskeletal remodeling and oxidative stress, impaired NPC migration, differentiation, and mitochondrial membrane potential, and increased ROS levels |
| [84] | Mitochondria transfer | Improved mitochondrial function and glutamatergic differentiation following mitochondria transfer |
| [85] | Expression studies in iNs, electrophysiology | miR-137 downregulates presynaptic plasticity genes and vesicle secretion in iNs |
| [55] | ATAC-seq, gene editing, SCS | Modification to a non-risk allele increases MIR137 expression and reduces neuronal maturation |
| [86] | Nanostring, RNA-seq, mass spectrometry | Reduced miR-9 expression inhibits outgrowth migration of NPC neurospheres |
| [87] | RNA-Seq, small RNA-seq, ChIP-Seq | Increased expression of miRNA networks, impaired miRNA dependent gene regulation |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, R.; Sportelli, V.; Ziller, M.; Spengler, D.; Hoffmann, A. Tracing Early Neurodevelopment in Schizophrenia with Induced Pluripotent Stem Cells. Cells 2018, 7, 140. https://0-doi-org.brum.beds.ac.uk/10.3390/cells7090140
Ahmad R, Sportelli V, Ziller M, Spengler D, Hoffmann A. Tracing Early Neurodevelopment in Schizophrenia with Induced Pluripotent Stem Cells. Cells. 2018; 7(9):140. https://0-doi-org.brum.beds.ac.uk/10.3390/cells7090140
Chicago/Turabian StyleAhmad, Ruhel, Vincenza Sportelli, Michael Ziller, Dietmar Spengler, and Anke Hoffmann. 2018. "Tracing Early Neurodevelopment in Schizophrenia with Induced Pluripotent Stem Cells" Cells 7, no. 9: 140. https://0-doi-org.brum.beds.ac.uk/10.3390/cells7090140