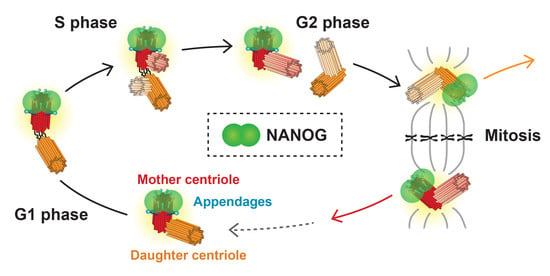
NANOG/NANOGP8 Localizes at the Centrosome and is Spatiotemporally Associated with Centriole Maturation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Immunofluorescence
2.3. Western Blotting
2.4. Transient Transfection
2.5. Immunoprecipitation and Protein Digestion
2.6. Mass Spectrometry and Data Processing
2.7. RT-qPCR
3. Results
3.1. Centrosomal Localization of the NANOG Protein
3.2. Validation of NANOG Centrosomal Signal Specificity and Anti-NANOG Antibodies
3.3. Analysis of NANOG and NANOGP8 Gene Expression
3.4. NANOG Colocalizes with the Mother Centriole
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chambers, I.; Colby, D.; Robertson, M.; Nichols, J.; Lee, S.; Tweedie, S.; Smith, A. Functional Expression Cloning of Nanog, a Pluripotency Sustaining Factor in Embryonic Stem Cells. Cell 2003, 113, 643–655. [Google Scholar] [CrossRef] [Green Version]
- Mitsui, K.; Tokuzawa, Y.; Itoh, H.; Segawa, K.; Murakami, M.; Takahashi, K.; Maruyama, M.; Maeda, M.; Yamanaka, S. The Homeoprotein Nanog Is Required for Maintenance of Pluripotency in Mouse Epiblast and ES Cells. Cell 2003, 113, 631–642. [Google Scholar] [CrossRef] [Green Version]
- Silva, J.; Nichols, J.; Theunissen, T.W.; Guo, G.; van Oosten, A.L.; Barrandon, O.; Wray, J.; Yamanaka, S.; Chambers, I.; Smith, A. Nanog is the gateway to the pluripotent ground state. Cell 2009, 138, 722–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hart, A.H.; Hartley, L.; Parker, K.; Ibrahim, M.; Looijenga, L.H.J.; Pauchnik, M.; Chow, C.W.; Robb, L. The pluripotency homeobox gene NANOG is expressed in human germ cell tumors. Cancer 2005, 104, 2092–2098. [Google Scholar] [CrossRef] [PubMed]
- Zbinden, M.; Duquet, A.; Lorente-Trigos, A.; Ngwabyt, S.-N.; Borges, I.; Ruiz i Altaba, A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 2010, 29, 2659–2674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagata, T.; Shimada, Y.; Sekine, S.; Hori, R.; Matsui, K.; Okumura, T.; Sawada, S.; Fukuoka, J.; Tsukada, K. Prognostic significance of NANOG and KLF4 for breast cancer. Breast Cancer 2014, 21, 96–101. [Google Scholar] [CrossRef]
- Santaliz-Ruiz, L.E.; Xie, X.; Old, M.; Teknos, T.N.; Pan, Q. Emerging Role of Nanog in Tumorigenesis and Cancer Stem Cells. Int. J. Cancer J. Int. Cancer 2014, 135, 2741–2748. [Google Scholar] [CrossRef]
- Fan, Z.; Li, M.; Chen, X.; Wang, J.; Liang, X.; Wang, H.; Wang, Z.; Cheng, B.; Xia, J. Prognostic Value of Cancer Stem Cell Markers in Head and Neck Squamous Cell Carcinoma: A Meta-analysis. Sci. Rep. 2017, 7, 43008. [Google Scholar] [CrossRef] [Green Version]
- Freitag, D.; McLean, A.L.; Simon, M.; Koch, A.; Grube, S.; Walter, J.; Kalff, R.; Ewald, C. NANOG overexpression and its correlation with stem cell and differentiation markers in meningiomas of different WHO grades. Mol. Carcinog. 2017, 56, 1953–1964. [Google Scholar] [CrossRef]
- Rasti, A.; Mehrazma, M.; Madjd, Z.; Abolhasani, M.; Zanjani, L.S.; Asgari, M. Co-expression of Cancer Stem Cell Markers OCT4 and NANOG Predicts Poor Prognosis in Renal Cell Carcinomas. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Nichols, J.; Zevnik, B.; Anastassiadis, K.; Niwa, H.; Klewe-Nebenius, D.; Chambers, I.; Schöler, H.; Smith, A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 1998, 95, 379–391. [Google Scholar] [CrossRef] [Green Version]
- Avilion, A.A.; Nicolis, S.K.; Pevny, L.H.; Perez, L.; Vivian, N.; Lovell-Badge, R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003, 17, 126–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, G.; Thomson, J.A. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007, 17, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Hyslop, L.; Stojkovic, M.; Armstrong, L.; Walter, T.; Stojkovic, P.; Przyborski, S.; Herbert, M.; Murdoch, A.; Strachan, T.; Lako, M. Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells Dayt. Ohio 2005, 23, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Schorle, H.; Nettersheim, D. NANOG (Nanog homeobox). Atlas Genet. Cytogenet. Oncol. Haematol. 2012. [Google Scholar] [CrossRef] [Green Version]
- Booth, H.A.F.; Holland, P.W.H. Eleven daughters of NANOG. Genomics 2004, 84, 229–238. [Google Scholar] [CrossRef]
- Palla, A.R.; Piazzolla, D.; Abad, M.; Li, H.; Dominguez, O.; Schonthaler, H.B.; Wagner, E.F.; Serrano, M. Reprogramming activity of NANOGP8, a NANOG family member widely expressed in cancer. Oncogene 2014, 33, 2513–2519. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Wang, X.; Li, M.; Han, J.; Chen, B.; Wang, B.; Dai, J. NANOGP8 is a retrogene expressed in cancers. FEBS J. 2006, 273, 1723–1730. [Google Scholar] [CrossRef]
- Ambady, S.; Malcuit, C.; Kashpur, O.; Kole, D.; Holmes, W.F.; Hedblom, E.; Page, R.L.; Dominko, T. Expression of NANOG and NANOGP8 in a variety of undifferentiated and differentiated human cells. Int. J. Dev. Biol. 2010, 54, 1743–1754. [Google Scholar] [CrossRef] [Green Version]
- Do, H.-J.; Lim, H.-Y.; Kim, J.-H.; Song, H.; Chung, H.-M.; Kim, J.-H. An intact homeobox domain is required for complete nuclear localization of human Nanog. Biochem. Biophys. Res. Commun. 2007, 353, 770–775. [Google Scholar] [CrossRef]
- Chang, D.F.; Tsai, S.C.; Wang, X.C.; Xia, P.; Senadheera, D.; Lutzko, C. Molecular Characterization of the Human NANOG Protein. Stem Cells 2009, 27, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.-T.; Liu, S.-Y.; Zheng, P.-S. Cytoplasmic NANOG-Positive Stromal Cells Promote Human Cervical Cancer Progression. Am. J. Pathol. 2012, 181, 652–661. [Google Scholar] [CrossRef] [PubMed]
- van Schaijik, B.; Davis, P.F.; Wickremesekera, A.C.; Tan, S.T.; Itinteang, T. Subcellular localisation of the stem cell markers OCT4, SOX2, NANOG, KLF4 and c-MYC in cancer: A review. J. Clin. Pathol. 2018, 71, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, P.; Dvorakova, D.; Koskova, S.; Vodinska, M.; Najvirtova, M.; Krekac, D.; Hampl, A. Expression and Potential Role of Fibroblast Growth Factor 2 and Its Receptors in Human Embryonic Stem Cells. STEM CELLS 2005, 23, 1200–1211. [Google Scholar] [CrossRef]
- Sana, J.; Zambo, I.; Skoda, J.; Neradil, J.; Chlapek, P.; Hermanova, M.; Mudry, P.; Vasikova, A.; Zitterbart, K.; Hampl, A.; et al. CD133 Expression and Identification of CD133/nestin Positive Cells in Rhabdomyosarcomas and Rhabdomyosarcoma Cell Lines. Anal. Cell. Pathol. Amst. 2011, 34, 303–318. [Google Scholar] [CrossRef]
- Loja, T.; Chlapek, P.; Kuglik, P.; Pesakova, M.; Oltova, A.; Cejpek, P.; Veselska, R. Characterization of a GM7 glioblastoma cell line showing CD133 positivity and both cytoplasmic and nuclear localization of nestin. Oncol. Rep. 2009, 21, 119–127. [Google Scholar]
- Veselska, R.; Skoda, J.; Loja, T.; Zitterbart, K.; Pavelka, Z.; Smardova, J.; Valaskova, I.; Hermanova, M.; Sterba, J. An unusual loss of EGFR gene copy in glioblastoma multiforme in a child: A case report and analysis of a successfully derived HGG-02 cell line. Childs Nerv. Syst. ChNS Off. J. Int. Soc. Pediatr. Neurosurg. 2010, 26, 841–846. [Google Scholar] [CrossRef]
- Veselska, R.; Janisch, R. Reaction of the Skin Fibroblast Cytoskeleton to Micromanipulation Interventions–ScienceDirect. Available online: https://0-www-sciencedirect-com.brum.beds.ac.uk/science/article/pii/S1047847701944326 (accessed on 2 August 2019).
- Mikulenkova, E.; Neradil, J.; Zitterbart, K.; Sterba, J.; Veselska, R. Overexpression of the ∆Np73 isoform is associated with centrosome amplification in brain tumor cell lines. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2015, 36, 7483–7491. [Google Scholar] [CrossRef]
- Kleylein-Sohn, J.; Westendorf, J.; Le Clech, M.; Habedanck, R.; Stierhof, Y.-D.; Nigg, E.A. Plk4-induced centriole biogenesis in human cells. Dev. Cell 2007, 13, 190–202. [Google Scholar] [CrossRef] [Green Version]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Holman, J.D.; Tabb, D.L.; Mallick, P. Employing ProteoWizard to Convert Raw Mass Spectrometry Data. Curr. Protoc. Bioinforma. 2014, 46, 13–24. [Google Scholar] [CrossRef] [PubMed]
- McIlwain, S.; Tamura, K.; Kertesz-Farkas, A.; Grant, C.E.; Diament, B.; Frewen, B.; Howbert, J.J.; Hoopmann, M.R.; Käll, L.; Eng, J.K.; et al. Crux: Rapid open source protein tandem mass spectrometry analysis. J. Proteome Res. 2014, 13, 4488–4491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The, M.; MacCoss, M.J.; Noble, W.S.; Käll, L. Fast and Accurate Protein False Discovery Rates on Large-Scale Proteomics Data Sets with Percolator 3.0. J. Am. Soc. Mass Spectrom. 2016, 27, 1719–1727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millikin, R.J.; Solntsev, S.K.; Shortreed, M.R.; Smith, L.M. Ultrafast Peptide Label-Free Quantification with FlashLFQ. J. Proteome Res. 2018, 17, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.C.; Gorenstein, M.V.; Li, G.-Z.; Vissers, J.P.C.; Geromanos, S.J. Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol. Cell. Proteomics MCP 2006, 5, 144–156. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods San Diego Calif 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ma, X.; Wang, B.; Wang, X.; Luo, Y.; Fan, W. NANOGP8 is the key regulator of stemness, EMT, Wnt pathway, chemoresistance, and other malignant phenotypes in gastric cancer cells. PLoS ONE 2018, 13, e0192436. [Google Scholar] [CrossRef] [Green Version]
- Hu, Q.; Khanna, P.; Ee Wong, B.S.; Lin Heng, Z.S.; Subhramanyam, C.S.; Thanga, L.Z.; Sing Tan, S.W.; Baeg, G.H. Oxidative stress promotes exit from the stem cell state and spontaneous neuronal differentiation. Oncotarget 2018, 9, 4223–4238. [Google Scholar] [CrossRef] [Green Version]
- Paintrand, M.; Moudjou, M.; Delacroix, H.; Bornens, M. Centrosome organization and centriole architecture: Their sensitivity to divalent cations. J. Struct. Biol. 1992, 108, 107–128. [Google Scholar] [CrossRef]
- Sonnen, K.F.; Schermelleh, L.; Leonhardt, H.; Nigg, E.A. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open 2012, 1, 965–976. [Google Scholar] [CrossRef] [Green Version]
- Nigg, E.A.; Holland, A.J. Once and only once: Mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Uzbekov, R.; Alieva, I. Who are you, subdistal appendages of centriole? Open Biol. 2018, 8, 180062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanos, B.E.; Yang, H.-J.; Soni, R.; Wang, W.-J.; Macaluso, F.P.; Asara, J.M.; Tsou, M.-F.B. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 2013, 27, 163–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vorobjev, I.A.; Nadezhdina, E.S. The centrosome and its role in the organization of microtubules. Int. Rev. Cytol. 1987, 106, 227–293. [Google Scholar] [PubMed]
- Bornens, M. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 2002, 14, 25–34. [Google Scholar] [CrossRef]
- Hinchcliffe, E.H. “It Takes Two to Tango”: Understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 2001, 15, 1167–1181. [Google Scholar] [CrossRef] [Green Version]
- Loncarek, J.; Khodjakov, A. Ab ovo or de novo? Mechanisms of centriole duplication. Mol. Cells 2009, 27, 135–142. [Google Scholar] [CrossRef] [Green Version]
- Fujita, H.; Yoshino, Y.; Chiba, N. Regulation of the centrosome cycle. Mol. Cell. Oncol. 2015, 3, e1075643. [Google Scholar] [CrossRef] [Green Version]
- Loncarek, J.; Bettencourt-Dias, M. Building the right centriole for each cell type. J. Cell Biol. 2018, 217, 823–835. [Google Scholar] [CrossRef]
- Stearns, T. Centrosome duplication. A centriolar pas de deux. Cell 2001, 105, 417–420. [Google Scholar] [CrossRef] [Green Version]
- Reina, J.; Gonzalez, C. When fate follows age: Unequal centrosomes in asymmetric cell division. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vorobjev, I.A.; Chentsov, Y.S. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 1982, 93, 938–949. [Google Scholar] [CrossRef] [PubMed]
- Piel, M.; Meyer, P.; Khodjakov, A.; Rieder, C.L.; Bornens, M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 2000, 149, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, M.M.; Malik, A.; Piel, M.; Bouckson-Castaing, V.; Bornens, M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: The role of ninein. J. Cell Sci. 2000, 113, 3013–3023. [Google Scholar] [PubMed]
- Ou, Y.Y.; Mack, G.J.; Zhang, M.; Rattner, J.B. CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J. Cell Sci. 2002, 115, 1825–1835. [Google Scholar]
- Lee, M.; Rhee, K. Determination of Mother Centriole Maturation in CPAP-Depleted Cells Using the Ninein Antibody. Endocrinol. Metab. Seoul Korea 2015, 30, 53–57. [Google Scholar] [CrossRef]
- Tateishi, K.; Yamazaki, Y.; Nishida, T.; Watanabe, S.; Kunimoto, K.; Ishikawa, H.; Tsukita, S. Two appendages homologous between basal bodies and centrioles are formed using distinct Odf2 domains. J. Cell Biol. 2013, 203, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Huang, N.; Xia, Y.; Zhang, D.; Wang, S.; Bao, Y.; He, R.; Teng, J.; Chen, J. Hierarchical assembly of centriole subdistal appendages via centrosome binding proteins CCDC120 and CCDC68. Nat. Commun. 2017, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Kashihara, H.; Chiba, S.; Kanno, S.; Suzuki, K.; Yano, T.; Tsukita, S. Cep128 associates with Odf2 to form the subdistal appendage of the centriole. Genes Cells 2019, 24, 231–243. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Glover, D.M. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012, 2, 120104. [Google Scholar] [CrossRef] [Green Version]
- Wilson, P.G.; Payne, T. Genetic reprogramming of human amniotic cells with episomal vectors: Neural rosettes as sentinels in candidate selection for validation assays. PeerJ 2014, 2, e668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuang, L.S.H.; Lai, S.K.; Murata-Hori, M.; Yamada, A.; Li, H.-Y.; Gunaratne, J.; Ito, Y. RUNX3 interactome reveals novel centrosomal targeting of RUNX family of transcription factors. Cell Cycle Georget. Tex 2012, 11, 1938–1947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madarampalli, B.; Yuan, Y.; Liu, D.; Lengel, K.; Xu, Y.; Li, G.; Yang, J.; Liu, X.; Lu, Z.; Liu, D.X. ATF5 Connects the Pericentriolar Materials to the Proximal End of the Mother Centriole. Cell 2015, 162, 580–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uzbekov, R.E. Centriole duplication in PE (SPEV) cells starts before the beginning of the DNA replication. Biochem. Mosc. Suppl. Ser. Membr. Cell Biol. 2007, 1, 206–211. [Google Scholar] [CrossRef]
- Gupta, A.; Tsuchiya, Y.; Ohta, M.; Shiratsuchi, G.; Kitagawa, D. NEK7 is required for G1 progression and procentriole formation. Mol. Biol. Cell 2017, 28, 2123–2134. [Google Scholar] [CrossRef]
- Singla, V.; Romaguera-Ros, M.; Garcia-Verdugo, J.M.; Reiter, J.F. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev. Cell 2010, 18, 410–424. [Google Scholar] [CrossRef] [Green Version]
- Ye, X.; Zeng, H.; Ning, G.; Reiter, J.F.; Liu, A. C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc. Natl. Acad. Sci. USA 2014, 111, 2164–2169. [Google Scholar] [CrossRef] [Green Version]
- Galati, D.F.; Mitchell, B.J.; Pearson, C.G. Subdistal Appendages Stabilize the Ups and Downs of Ciliary Life. Dev. Cell 2016, 39, 387–389. [Google Scholar] [CrossRef] [Green Version]
- Mazo, G.; Soplop, N.; Wang, W.-J.; Uryu, K.; Tsou, M.-F.B. Spatial Control of Primary Ciliogenesis by Subdistal Appendages Alters Sensation-Associated Properties of Cilia. Dev. Cell 2016, 39, 424–437. [Google Scholar] [CrossRef] [Green Version]
- Chang, P.; Giddings, T.H.; Winey, M.; Stearns, T. Epsilon-tubulin is required for centriole duplication and microtubule organization. Nat. Cell Biol. 2003, 5, 71–76. [Google Scholar] [CrossRef]
- Yamashita, Y.M.; Mahowald, A.P.; Perlin, J.R.; Fuller, M.T. Asymmetric Inheritance of Mother Versus Daughter Centrosome in Stem Cell Division. Science 2007, 315, 518–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelletier, L.; Yamashita, Y.M. Centrosome asymmetry and inheritance during animal development. Curr. Opin. Cell Biol. 2012, 24, 541–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, Y.M.; Jones, D.L.; Fuller, M.T. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 2003, 301, 1547–1550. [Google Scholar] [CrossRef] [PubMed]










| Cell Line | Tissue Type | Source |
|---|---|---|
| Tumor Cell Lines | ||
| RD | Embryonal rhabdomyosarcoma | ECACC, Cat. No. 85111502 |
| NSTS-11 | Rhabdomyosarcoma | [25] |
| NSTS-34 | Rhabdomyosarcoma | Established in the laboratory |
| NSTS-35 | Rhabdomyosarcoma | Established in the laboratory |
| GM7 | Glioblastoma multiforme | [26] |
| HGG-02 | Glioblastoma multiforme | [27] |
| Daoy | Medulloblastoma | ATCC, HTB-186TM |
| SH-SY5Y | Neuroblastoma | ECACC, Cat. No. 94030304 |
| NTERA-2 | Embryonal carcinoma | ECACC, Cat. No. 01071221 |
| Non-Tumor Cell Lines | ||
| CCTL14 | Human embryonal stem cells | [24] |
| KF1 | Normal fibroblasts | [28] |
| Primary Antibodies | |||||
|---|---|---|---|---|---|
| Antigen | Isotype | Clone (Cat. No.) | Manufacturer | Dilution | |
| IF | WB | ||||
| Nanog | Rb IgG | D73G4 (4903) | CST | 1:200 | 1:2000 |
| Nanog | Rb IgG | EPR2027(2) (ab109250) | Abcam | 1:200 | 1:2000 |
| Pericentrin | Rb IgG | - (ab4448) | Abcam | 1:400 | - |
| CP110 | Mo | - | - | 1:5 | - |
| Ninein | Mo IgG2a | 79.160-7 (41-3400) | Thermo Fisher | 1:800 | - |
| α-Tubulin | Mo IgG1 | DM1A (ab7291) | Abcam | - | 1:10,000 |
| Lamin B2 | Rb IgG | D8P3U (12255) | CST | - | 1:400 |
| Secondary Antibodies | |||||
| Host | Specificity | Conjugate | Manufacturer | Dilution | |
| IF | WB | ||||
| Donkey | anti-Mo IgG | Alexa Fluor 488 | Life Technologies | 1:200 | - |
| Donkey | anti-Rb IgG | Alexa Fluor 488 | Life Technologies | 1:200 | - |
| Donkey | anti-Mo IgG | Alexa Fluor 568 | Life Technologies | 1:200 | - |
| Donkey | anti-Rb IgG | Alexa Fluor 568 | Life Technologies | 1:200 | - |
| Horse | anti-Mo IgG | HRP | CST | - | 1:5000 |
| Goat | anti-Rb IgG | HRP | CST | - | 1:5000 |
| Gene | Gene Symbol | Primer Sequence |
|---|---|---|
| Nanog homeobox | NANOG | F: 5′-TTCATTATAAATCTAGAGACTCCAGGA-3′ |
| R: 5′-CTTTGGGACTGGTGGAAGAATC-3′ | ||
| Nanog homeobox/Nanog homeobox retrogene P8 * | NANOG/P8 | F: 5′-GCAGAGAAGAGTGTCG-3′ |
| R: 5′-AGCTGGGTGGAAGAGAACACAG-3′ | ||
| Heat shock protein 90 alpha family class B member 1 | HSP90AB1 | F: 5′-CGCATGAAGGAGACACAGAA-3′ |
| R: 5′-TCCCATCAAATTCCTTGAGC-3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mikulenkova, E.; Neradil, J.; Vymazal, O.; Skoda, J.; Veselska, R. NANOG/NANOGP8 Localizes at the Centrosome and is Spatiotemporally Associated with Centriole Maturation. Cells 2020, 9, 692. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9030692
Mikulenkova E, Neradil J, Vymazal O, Skoda J, Veselska R. NANOG/NANOGP8 Localizes at the Centrosome and is Spatiotemporally Associated with Centriole Maturation. Cells. 2020; 9(3):692. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9030692
Chicago/Turabian StyleMikulenkova, Erika, Jakub Neradil, Ondrej Vymazal, Jan Skoda, and Renata Veselska. 2020. "NANOG/NANOGP8 Localizes at the Centrosome and is Spatiotemporally Associated with Centriole Maturation" Cells 9, no. 3: 692. https://0-doi-org.brum.beds.ac.uk/10.3390/cells9030692